Back to article: Sulfur transfer and activation by ubiquitin-like modifier system Uba4•Urm1 link protein urmylation and tRNA thiolation in yeast
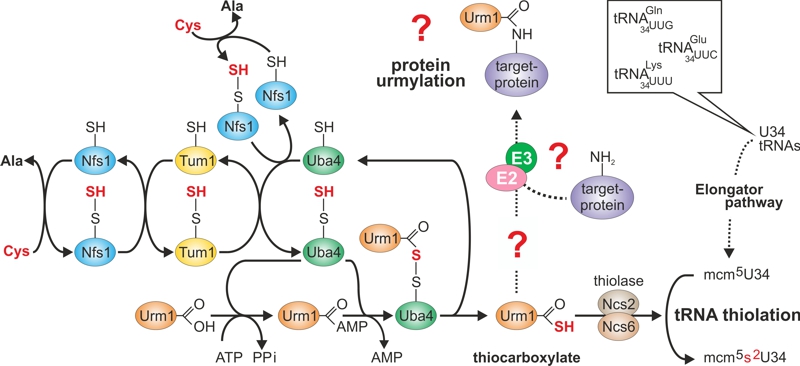
FIGURE 1: Sulfur flow within the URM1 pathway.
The scheme depicts sulfur (red) flow and URM1 pathway players required for mobilization (Nfs1), transfer (Tum1), activation (Uba4, Urm1) or consumption (Ncs2•Ncs6) of sulfur. E1-like activator Uba4 (green) is key to Urm1 thiocarboxylation. Urm1-COSH formed this way donates sulfur to the tRNA thiolation branch which cooperates with the Elongator pathway to form 5-methoxy-carbonyl-methyl-2-thio-uridine in wobble positions (mcm5s2U34) of the indicated tRNA anticodons. Possibly (?) Urm1-COSH also feeds into the urmylation branch of the URM1 pathway. As for the latter, E2/E3 enzymes are elusive (?) and the relevance of urmylation for target protein function is ill-defined (?). The model is up-dated from work in the labs of Hayashi [7] and Suzuki [9].
7. Nakai Y, Nakai M, and Hayashi H (2008). Thio-modification of yeast cytosolic tRNA requires a ubiquitin-related system that resembles bacterial sulfur transfer systems. J Biol Chem 283(41): 27469-27476. https://doi.org/10.1111/1567-1364.12039
9. Noma A, Sakaguchi Y, and Suzuki T (2009). Mechanistic characteriza-tion of the sulfur-relay system for eukaryotic 2-thiouridine biogenesis at tRNA wobble positions. Nucleic Acids Res 37(4): 1335-1352. https://doi.org/10.1093/nar/gkn1023