Back to article: The copper transport-associated protein Ctr4 can form prion-like epigenetic determinants in Schizosaccharomyces pombe
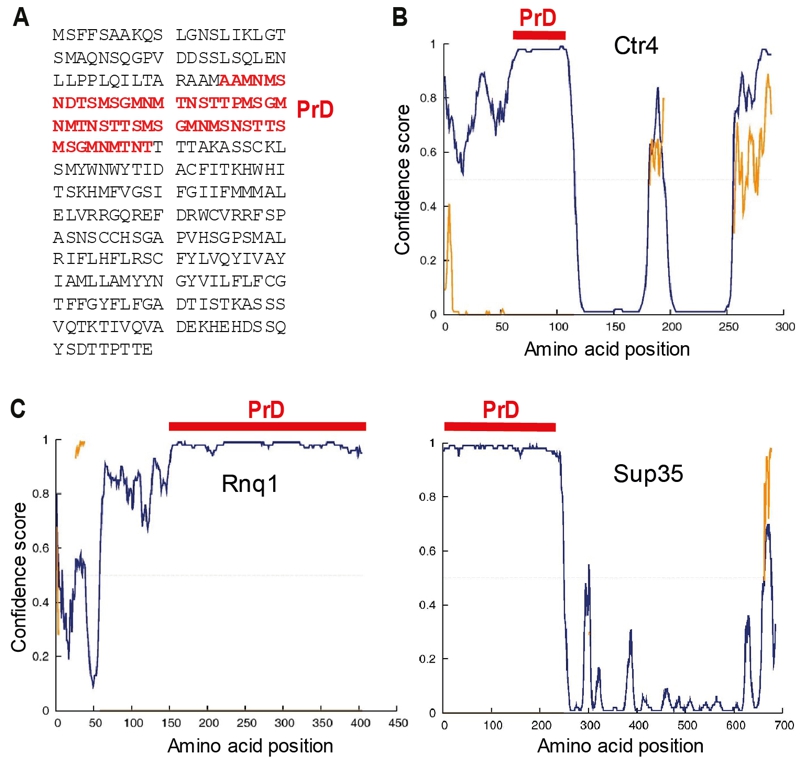
FIGURE 2: Sequence features of Ctr4.
(A) The 289 amino acid Ctr4 protein contains a 55 amino acid prion-forming domain (PrD, red) as predicted by the PLAAC algorithm [49].
(B) The predicted PrD of Ctr4 (red bar) coincides with the highest predicted unfolded region (disordered, blue curve) according to the DISOPRED3 algorithm [51]. The yellow trace is the location of predicted protein binding sites within disordered regions.
(C) DISOPRED3 predictions of intrinsically disordered regions in two prion-forming proteins of S. cerevisiae, Rnq1 (left) and Sup35 (right), together with the locations of the experimentally defined PrDs, as in (B).
49. Lancaster AK, Nutter-Upham A, Lindquist S, King OD (2014). PLAAC: a web and command-line application to identify proteins with prion-like amino acid composition. Bioinformatics 30(17): 2501-2502. https://doi.org/10.1093/bioinformatics/btu310
51. Buchan DW, Minneci F, Nugent TC, Bryson K, Jones DT (2013). Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res 41(Web Server issue): W349-357. https://doi.org/10.1093/nar/gkt381