Back to article: Identification of SUMO conjugation sites in the budding yeast proteome
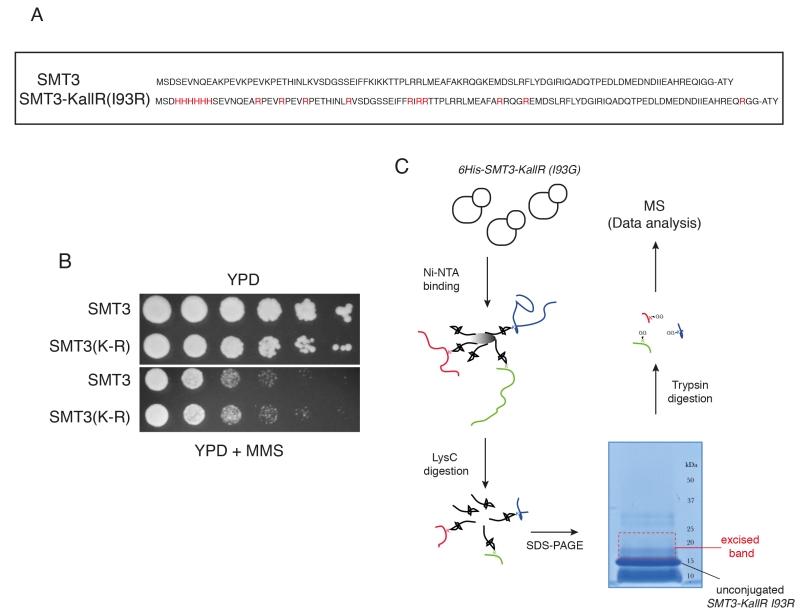
FIGURE 1: Proteomic screen to identify SUMO sites in budding yeast proteins. (A) Sequences of wildtype SUMO (SMT3) and the lysine-deficient His6 tagged mutant (SMT3-KallR I93R) used in the method. (B) Wild-type and SMT3-KallR I93R strains plated on full media (YPD) and media containing methyl methanesulphonate (MMS). (C) Diagrammatic representation of the purification strategy employed to enrich for SMT3-conjugated peptides. Cell lysates from yeast expressing SMT3-KallR I93R were digested with the endoprotease LysC, cleaving after lysines. The digested lysates were run on SDS-PAGE. Unconjugated SMT3-KallR I93R (indicated) and SMT3-KallR I93R conjugated to target protein fragments were identified. Gel area above the unconjugated SMT3-KallR I93R band were excised, digested with trypsin, and analyzed by nano LC-MS/MS. And database searched to identify SUMO acceptor lysines in the yeast proteome.