Back to article: Identification of SUMO conjugation sites in the budding yeast proteome
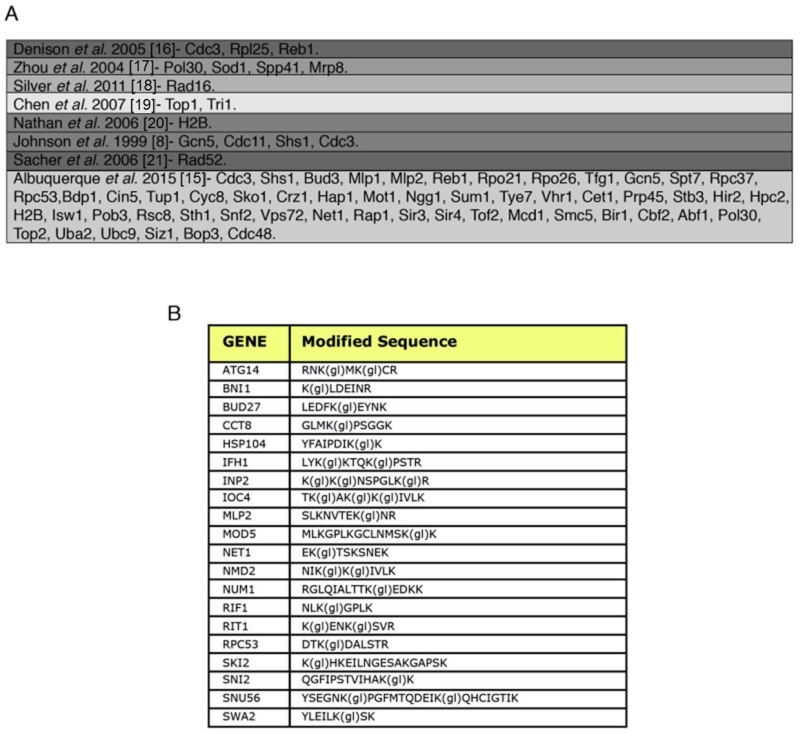
FIGURE 2: SUMO-acceptor lysines detected in our mass spectrometry analysis were compared with SUMO-acceptor lysines already described in previous studies done in S. cerevisiae. (A) Previous studies were based on either MS or site-directed mutagenesis/immunoblotting, or a combination of both. Most of the SUMO-acceptor lysines previously found were also detected in this study. SUMO substrates (total of 257) identified in the yeast proteome of cells grown asynchronously. Previously published SUMO-substrates were obtained [8,15-21]. (B) To ensure that diglycine-modified lysines detected by mass spectrometry after Smt3 purification are not due to modification by ubiquitin or Nedd8, a centromeric plasmid 8His-SMT3-KallR-REQIGG-pRS415 expressing the Smt3 variant used previously for the Smt3 purification protocol with the difference that the RGG conjugating terminus was replace for the native RIEQGG C-terminus was employed. SUMO-acceptor lysines modified by the 8His-Smt3-KallR-REQIGG keep a side chain of 5 aa after trypsin digestion (EQIGG). Therefore, any diglycine-modified lysines detected by mass spectrometry under these conditions can only be due to either false positive hits, or to ubiquitinated or neddylated contaminants. A large culture of 9 l of the strain expressing the 8His-SMT3-KallR-REQIGG variant was grown in YPD and harvested at O.D. 0.9. Smt3 purification and mass spectrometry analysis was performed as described in Material and Methods. We detected 23 diglycine-modified lysines. None of these corresponded with previously detected diglycine-modified lysines in our Smt3-RGG pulldowns. In addition none of these diglycine-modified lysines are within SUMO consensus sequences. This strongly indicates that sumo-acceptor lysines identified after purification of Smt3-RGG pulldowns represent bona fide SUMOylation sites.
8. Johnson ES, Blobel G (1999). Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. (J Cell Biol 147(5): 981-994. https://www.ncbi.nlm.nih.gov/pubmed/?term=10579719
15. Albuquerque CP, Yeung E, Ma S, Fu T, Corbett KD, Zhou H (2015). A Chemical and Enzymatic Approach to Study Site-Specific Sumoylation. (PLoS One 10(12): e0143810. https://doi.org/10.1371/journal.pone.0143810
16. Denison C, Rudner AD, Gerber SA, Bakalarski CE, Moazed D, Gygi SP (2005). A proteomic strategy for gaining insights into protein sumoylation in yeast. (Mol Cell Proteomics 4(3): 246-254. https://doi.org/10.1074/mcp.M400154-MCP200
17. Zhou W, Ryan JJ, Zhou H (2004). Global analyses of sumoylated proteins in Saccharomyces cerevisiae. Induction of protein sumoylation by cellular stresses. ( J Biol Chem 279(31): 32262-32268. https://doi.org/10.1074/jbc.M404173200
18. Silver HR, Nissley JA, Reed SH, Hou YM, Johnson ES (2011). A role for SUMO in nucleotide excision repair. (DNA Repair (Amst) 10(12): 1243-1251. https://doi.org/10.1016/j.dnarep.2011.09.013
19. Chen XL, Silver HR, Xiong L, Belichenko I, Adegite C, Johnson ES (2007). Topoisomerase I-dependent viability loss in Saccharomyces cerevisiae mutants defective in both SUMO conjugation and DNA repair. (Genetics 177(1): 17-30. https://doi.org/10.1534/genetics.107.074708
20. Nathan D, Ingvarsdottir K, Sterner DE, Bylebyl GR, Dokmanovic M, Dorsey JA, Whelan KA, Krsmanovic M, Lane WS, Meluh PB, Johnson ES, Berger SL (2006). Histone sumoylation is a negative regulator in Saccharomyces cerevisiae and shows dynamic interplay with positive-acting histone modifications. (Genes Dev 20(8): 966-976. https://doi.org/10.1101/gad.1404206
21. Sacher M, Pfander B, Hoege C, Jentsch S (2006). Control of Rad52 recombination activity by double-strand break-induced SUMO modification. (Nat Cell Biol 8(11): 1284-1290. https://doi.org/10.1038/ncb1488