Back to article: Cross-species complementation of bacterial- and eukaryotic-type cardiolipin synthases
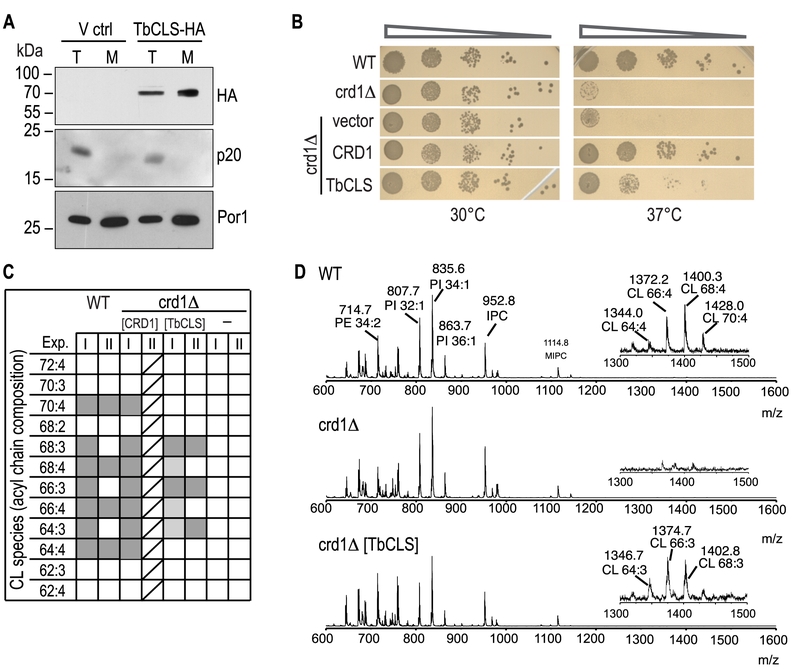
FIGURE 2: Expression, localization, and functionality of TbCLS in a S. cerevisiae crd1Δ background. (A) SDS-PAGE/immunoblot analysis using anti-HA antibodies of cell lysates (T) and mitochondria (M) prepared from crd1Δ cells transfected with the empty expression vector (V ctrl) or Su9-TbCLS-HA (TbCLS-HA). Antibodies against the cytosolic protein p20 and mitochondrial porin Por1 were used as controls. (B) Growth and colony formation in serial dilutions of S. cerevisiae wild-type (WT), crd1Δ, and crd1Δ cells expressing Su9-TbCLS, Crd1, or the empty expression vector. Plates were incubated in parallel at 30°C and 37°C for 4 days. (C) Cardiolipin (CL) species composition of wild-type (WT), crd1Δ (-), Crd1 addback (CRD1), and TbCLS complemented crd1Δ cells (TbCLS). Lipid extracts from crude mitochondrial membranes (Exp. I) or spheroplasts (Exp. II) were analyzed by MALDI-TOF mass spectrometry in two independent experiments. Molecular species are defined by the total number of carbon atoms in the acyl chains and the number of unsaturated bonds within the acyl chain. Identified species are marked in dark grey, minor species are in light gray; only one analysis was done with the CRD1 addback sample (indicated by diagonal lines). (D) Representative full-range mass spectra from experiment II; the insets highlight the m/z range of the cardiolipin molecular species of wild-type (WT, top panel) and TbCLS-complemented crd1Δ cells (crd1Δ [TbCLS], bottom panel); no cardiolipin species are detected in crd1Δ cells (middle panel).