Back to article: Guidelines for DNA recombination and repair studies: Cellular assays of DNA repair pathways
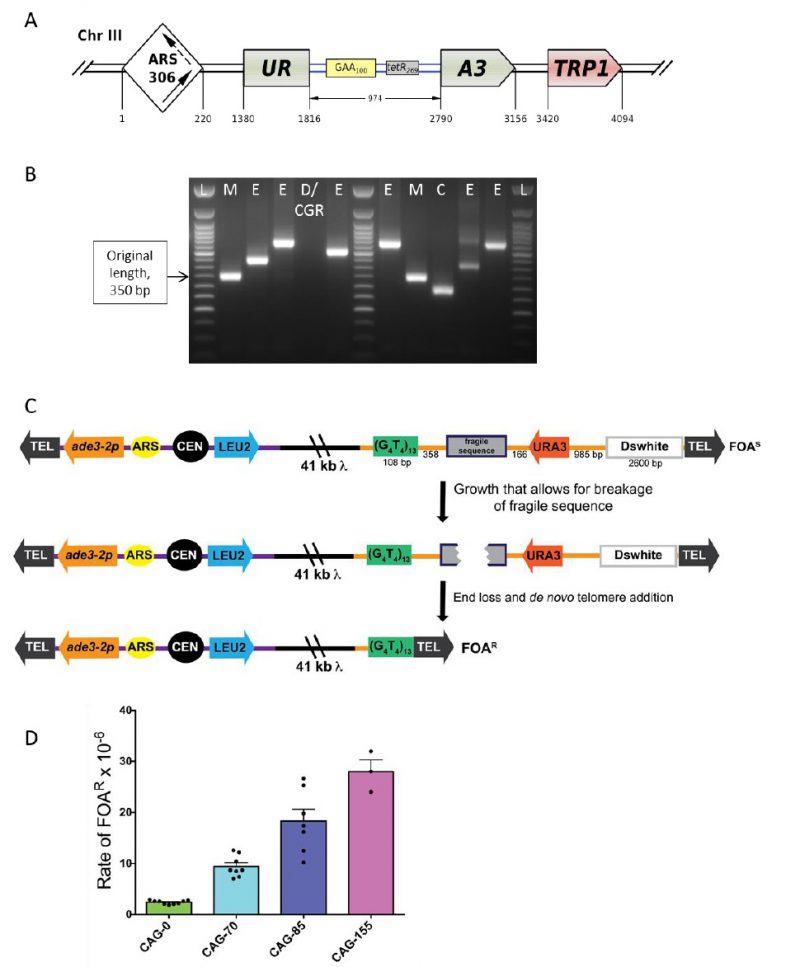
FIGURE 6: Scheme of the genetically tractable systems to study repeat expansions and repeat-mediated genome instability in yeast. (A) Scheme of the genetically tractable system to study repeat expansions and repeat-mediated genome instability in yeast. The selectable cassette contains flanking sequences from chromosome III, the URA3 gene from the pYES2 plasmid (Invitrogen) (green), an intron sequence derived from the ACT1 gene from chromosome VI (inside the URA3 gene) (blue), part of the tetR coding region from the pACY184 plasmid (NEB) (gray), and the TRP1 gene from the pYES3/CT plasmid (Invitrogen) (red). (B) Gel electrophoresis of the PCR analysis of 5-FOAr colonies derived from a fluctuation test of a strain with 100 GAA repeats. L – ladder, M – mutation, E – expansion, D/CGR – deletion or CGR (complex genomic rearrangements), C – contraction with simultaneous repeat-mediated mutagenesis. The black arrow indicates a ∼350bp PCR fragment of the original (GAA)100 repeat. (C) Assay to measure chromosome fragility. In this system, a fragile sequence has been integrated onto a yeast artificial chromosome (YAC) between a telomere seed sequence (G4T4)13 and the URA3 gene. Breaks that occur within the fragile sequence are subject to resection and telomere addition at the G4T4 sequence, which results in loss of the URA3 gene and renders cells 5-FOAR. The YAC additionally contains a LEU2 marker gene, which allows for maintenance of the YAC, a centromere (CEN4), an origin of replication (ARS1), yeast telomeres (TEL), a partially functional ade3-2p allele, and the Drosophila white gene. The orange line indicates pYIP5 plasmid backbone, the black line indicates lambda DNA, and the purple line corresponds to pUC18 plasmid backbone. (D) Wild-type fragility data of S. cerevisiae strain BY4705 where the fragile sequence integrated between the G4T4 and URA3 marker is a CAG tract of the indicated length, in number of repeats. Assays done with this (CAG)n-URA3 YAC show that 5-FOA resistance increases with increasing number of CAG repeats. Data sourced from [107, 117, 365, 366, 367].
107. Kerrest A, Anand RP, Sundararajan R, Bermejo R, Liberi G, Dujon B, Freudenreich CH, Richard GF (2009). SRS2 and SGS1 prevent chromosomal breaks and stabilize triplet repeats by restraining recombination. Nat Struct Mol Biol 16(2): 159-167. doi: 10.1038/nsmb.1544
117. Sundararajan R, Gellon L, Zunder RM, Freudenreich CH (2010). Double-strand break repair pathways protect against CAG/CTG repeat expansions, contractions and repeat-mediated chromosomal fragility in Saccharomyces cerevisiae. Genetics 184(1): 65-77. doi: 10.1534/genetics.109.111039
365. House NC, Yang JH, Walsh SC, Moy JM, Freudenreich CH (2014). NuA4 Initiates Dynamic Histone H4 Acetylation to Promote High-Fidelity Sister Chromatid Recombination at Postreplication Gaps. Mol Cell 55(6): 818-828. doi: 10.1016/j.molcel.2014.07.007
366. Nguyen JH, Viterbo D, Anand RP, Verra L, Sloan L, Richard GF, Freudenreich CH (2017). Differential requirement of Srs2 helicase and Rad51 displacement activities in replication of hairpin-forming CAG/CTG repeats. Nucleic Acids Res 45(8):4519-4531. doi: 10.1093/nar/gkx088.
367. Yang JH, Freudenreich CH (2010). The Rtt109 histone acetyltransferase facilitates error-free replication to prevent CAG/CTG repeat contractions. DNA Repair 9(4): 414-420. doi: 10.1016/j.dnarep.2009.12.022