Back to article: Raman-based sorting of microbial cells to link functions to their genes
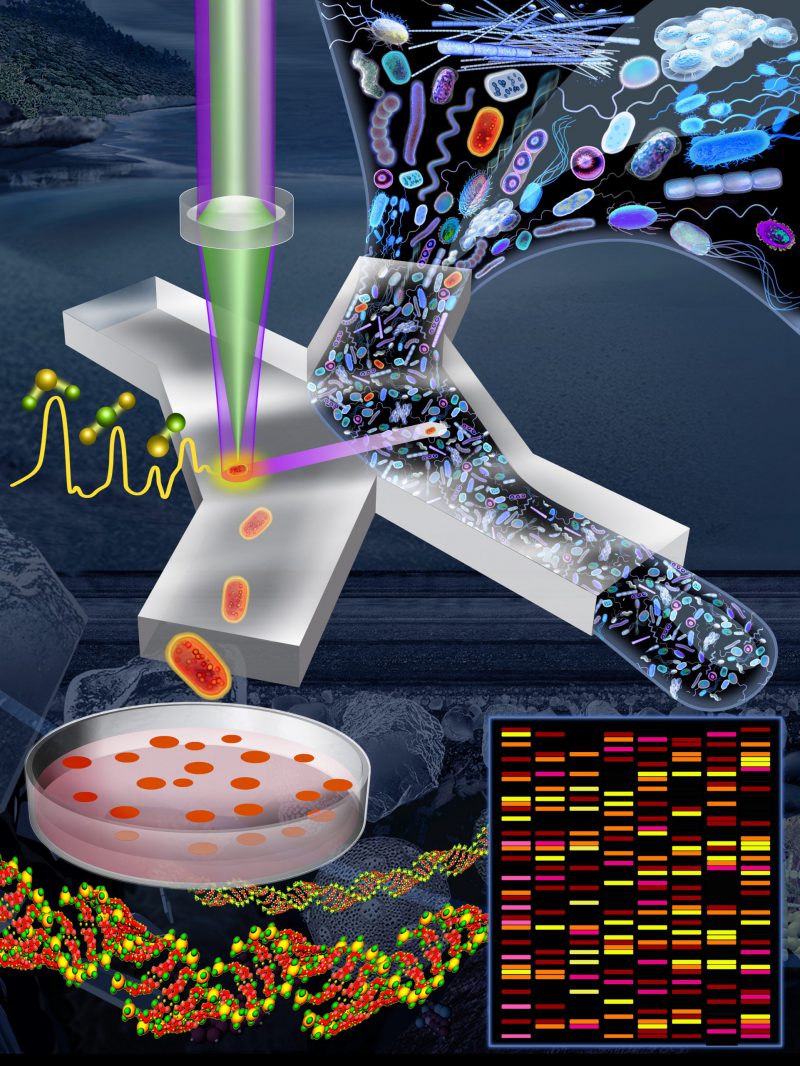
FIGURE 1: Pipeline to link functions of microbial cells to their genes. An environmental sample (e.g., marine, freshwater, soil, mammalian gut) is incubated for a short period in D2O-containing minimal medium (i.e., lacking nutrients) supplemented by a specific compound of interest (this initial step is not shown in this image). Cells able to metabolize that compound are active and so become labelled by deuterium (D), shown here in red. The sample is introduced into a microfluidic device in which single cells are captured in optical tweezers and analyzed using a Raman microspectroscope. The status of deuterium-labelled cells is apparent from a characteristic peak in their Raman spectrum. Deuterium-labelled cells (i.e., those with the metabolic property of interest) pass into a collection outlet and are available for subsequent genetic analysis (e.g., metagenomics, single-cell genomics) or cultivation for further phenotypic characterization. Image credit: Lee KS, Gorick G, and Stocker R.