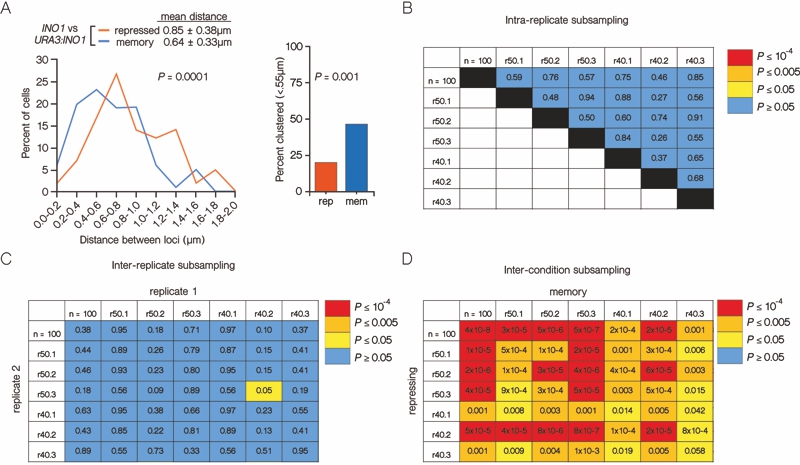
FIGURE 2: INO1 transcriptional memory leads to interchromosomal clustering.
(A) Haploid cells having the endogenous INO1 gene marked with the TetO and URA3:INO1 marked with the LacO, expressing GFP-TetR and mRFP-LacI [8] were grown under INO1 repressing (+ inositol) or memory (- inositol → + inositol, 3h) conditions, fixed and processed for immunofluorescence against GFP and mRFP. Left: The distribution of distances between the two loci in ~100 cells, binned into 0.2 µm bins. P values were calculated using a Wilcoxon Rank Sum Test. Right: the fraction of cells in which the two loci were ≤ 0.55 µm. P values were calculated using a Fisher Exact Test. Note: the distribution of the repressed condition has been previously published [7] and is shown only for comparison to the distribution under the experimental (memory) condition.
(B-D) Subsampling analysis. Full datasets (n = 100) or randomly generated subsamples of 50 or 40 measurements (r50 or r40, respectively) were compared pairwise using a Wilcoxon Rank Sum test. The numbers in each cell are the P values, color-coded as described in the legend.
(B) A biological replicate compared with itself.
(C) Two biological replicates compared with each other.
(D) Distributions from repressing and memory conditions compared with each other.
7. Brickner DG, Ahmed S, Meldi L, Thompson A, Light WH, Young M, Hickman TL, Chu F, Fabre E and Brickner JH (2012). Transcription factor binding to a DNA zip code controls interchromosomal clustering at the nuclear periphery. Dev Cell 22:1234-1246. http://dx.doi.org/10.1016/j.devcel.2012.03.012
8. Brickner DG, Cajigas I, Fondufe-Mittendorf Y, Ahmed S, Lee PC, Widom J and Brickner JH (2007). H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol 5:e81. http://dx.doi.org/10.1371/journal.pbio.0050081