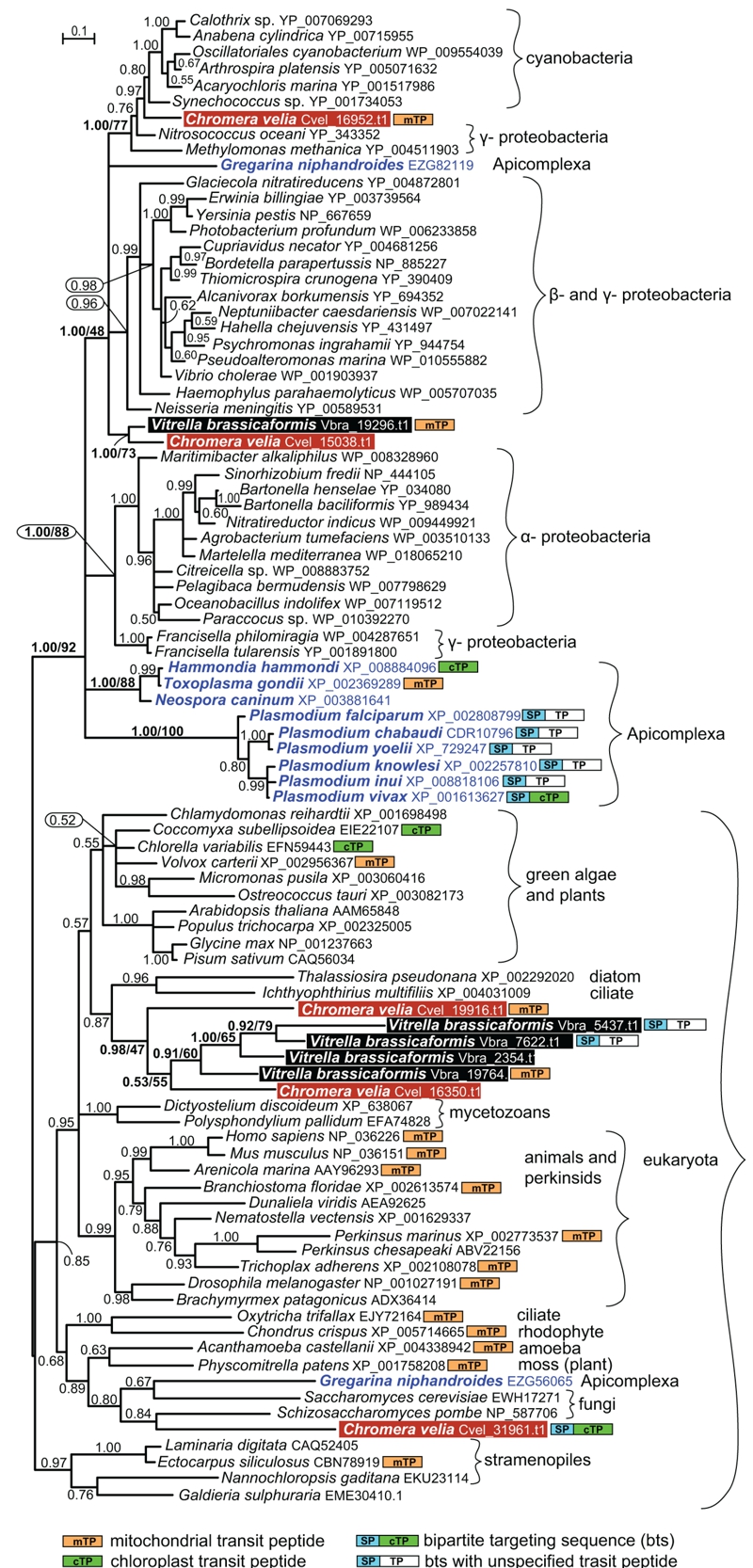
FIGURE 1: Bayesian phylogenetic tree as inferred from Prx5 amino acid sequences.
The original dataset used by Djuika et al. was extended by the addition of relevant sequences from the chromerid algae Chromera velia and Vitrella brassicaformis, and the early-branching apicomplexan Gregarina niphandroides. Sequences were aligned using MAFFT, edited by Gblocs (both implemented in SeaView), and the alignment was analyzed by Bayesian inference and maximum likelihood (ML) method. MrBayes 3.2 [20] was used to assess Bayesian topologies and posterior probabilities (PP) using WAG model. Two independent Monte-Carlo Markov chains were run under the default settings for 3 million generations; we used 500,000 generations as a burn-in and omitted them from topology reconstruction and PP calculation. ML bootstrap support is shown for selected nodes in the tree; ML tree was computed by RAxML [21], using γ corrected WAG model with bootstrap analysis being inferred from 1,000 replicates. Putative intracellular localizations of the Prx5 proteins were predicted using the TargetP and SignalP programs, respectively [22].
20. Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic biology 61(3): 539-542. http://dx.doi.org/10.1093/sysbio/sys029
21. Stamatakis A, Ludwig T, Meier H (2005). RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics 21(4): 456-463. http://dx.doi.org/10.1093/bioinformatics/bti191
22. Emanuelsson O, Brunak S, von Heijne G, Nielsen H (2007). Locating proteins in the cell using TargetP, SignalP and related tools. Nature protocols 2(4): 953-97. http://dx.doi.org/10.1038/nprot.2007.131