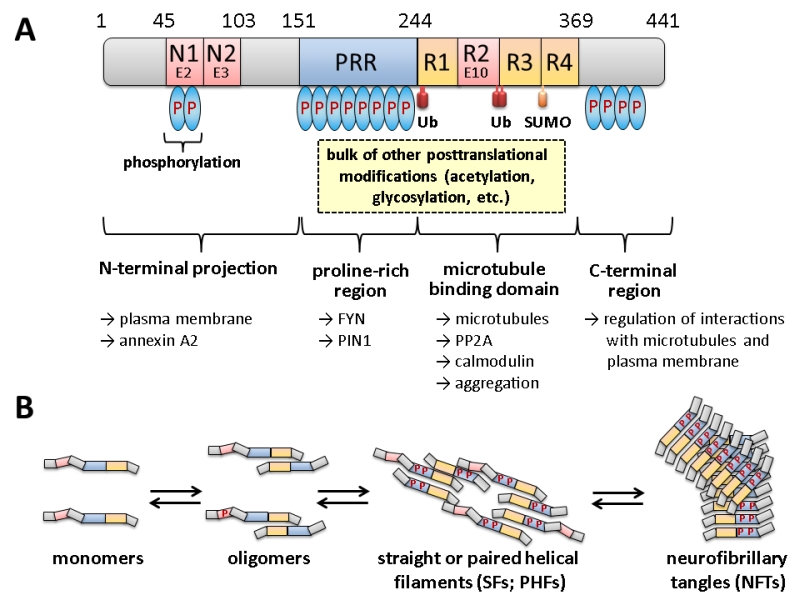
FIGURE 1: Domain structure and oligomerization of tau. (A) The longest isoform of human tau present in neurons of the central nervous system (CNS) is shown, which differs from other CNS isoforms in the presence or absence of two N-terminal domains (encoded by exons E2 and E3) and one of the repeats in the microtubule interaction domain (R2 encoded by exon E10). For variations in numbering and nomenclature consult [8]. Modifications by ubiquitination (Ub) and SUMOylation (SUMO) are indicated below. Not all phosphorylation sites (P in blue ovals) are shown, but the relative degrees are indicated by the number of ovals. Most of the other covalent modifications take place within the proline-rich region (PRR) and the R1-R4 motif. Some of the cellular structures and proteins for which interaction of the respective domains has been suggested are designated by the arrows shown below. This scheme summarizes informations from several excellent reviews, from which more detailed descriptions of the different modifications and their effect on tau in addition to those mentioned in the text can be obtained: [8][9][10][11][12]. (B) Different oligomerization states of tau are shown. It is believed that the monomeric form fulfills the physiological functions, while soluble oligomers and/or PHFs are responsible for the neurotoxic effects. As indicated, the formation of oligomers is accompanied by increasing degrees of phosphorylation at different sites, as well as by proteolytic trimming (adapted and simplified from [13]).
8. Bakota L, Brandt R (2016).Tau Biology and Tau-Directed Therapies for Alzheimer’s Disease. Drugs 76(3):301-13. http://dx.doi.org/10.1007/s40265-015-0529-0
9. De Vos A, Anandhakumar J, Van den Brande J, Verduyckt M, Franssens V, Winderickx J, Swinnen E (2011). Yeast as a model system to study tau biology. Int J Alzheimers Dis 2011:428970. http://dx.doi.org/10.4061/2011/428970
10. Fontaine SN, Sabbagh JJ, Baker J, Martinez-Licha CR, Darling A, Dickey CA (2015). Cellular factors modulating the mechanism of tau protein aggregation. Cell Mol Life Sci 72(10): 1863-1879. http://dx.doi.org/10.1007/s00018-015-1839-9
11. Ihara Y, Morishima-Kawashima M, Nixon R (2012). The ubiquitin-proteasome system and the autophagic-lysosomal system in Alzheimer disease. Cold Spring Harb Perspect Med 2(8). http://dx.doi.org/10.1101/cshperspect.a006361
12. Mandelkow EM, Mandelkow E (2012). Biochemistry and cell biology of tau protein in neurofibrillary degeneration. Cold Spring Harb Perspect Med 2(7): a006247. http://dx.doi.org/10.1101/cshperspect.a006247
13. Flores-Rodriguez P, Ontiveros-Torres MA, Cardenas-Aguayo MC, Luna-Arias JP, Meraz-Rios MA, Viramontes-Pintos A, Harrington CR, Wischik CM, Mena R, Floran-Garduno B, Luna-Munoz J (2015). The relationship between truncation and phosphorylation at the C-terminus of tau protein in the paired helical filaments of Alzheimer’s disease. Front Neurosci 9:33. http://dx.doi.org/10.3389/fnins.2015.00033