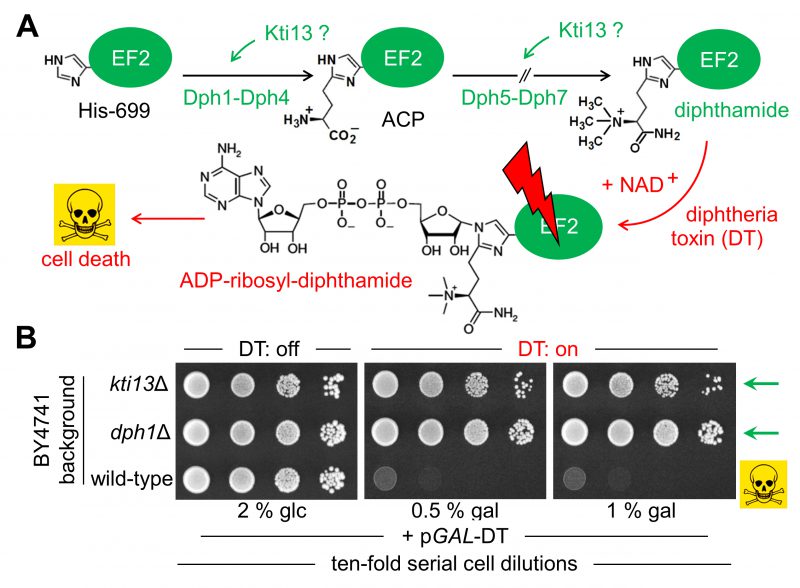
FIGURE 1: Potential role of yeast KTI13 in diphthamide modification. (A) Simplified pathway overview [24][25]. Diphthamide synthesis initiates with modifcation of EF2 at His-699 by ACP involving proteins Dph1-Dph4. Subsequential reactions to convert ACP into end product diphthamide entail Dph5-Dph7. Potential Kti13 involvement in the synthesis steps is indicated (‘?’). Diphthamide can be hijacked by diphtheria toxin (DT) for ADP-ribosylation in an NAD+ fashion and induces cell death by EF2 inactivation (skull-crossbones). (B) KTI13 and DPH1 gene deletion strains resist against DT cytotoxicity. Yeast strains carrying pGAL-DT [39], a plasmid for galactose-inducible expression of the lethal ADP-ribosylase domain from DT (see A) were spotted onto medium containing 0.5-1% (w/v) galactose (gal) or 2% (w/v) glucose (glc). Following DT induction, growth inhibition of diphthamide-proficient wild-type is distinguishable from DT resistance of diphthamide-deficient dph1Δ and kti13Δ mutants (green arrows).
By continuing to use the site, you agree to the use of cookies. more information
The cookie settings on this website are set to "allow cookies" to give you the best browsing experience possible. If you continue to use this website without changing your cookie settings or you click "Accept" below then you are consenting to this. Please refer to our "privacy statement" and our "terms of use" for further information.