Back to article: Guidelines for DNA recombination and repair studies: Cellular assays of DNA repair pathways
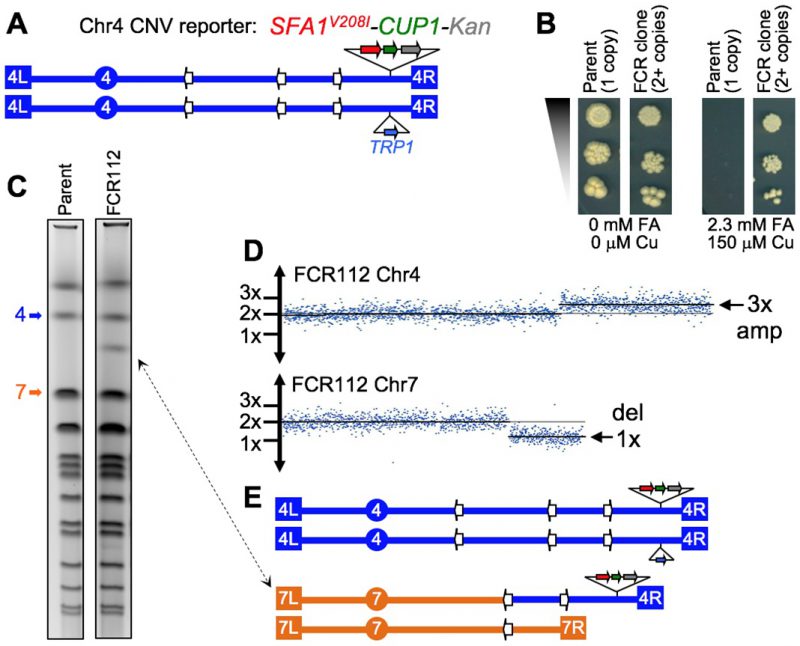
FIGURE 7: The diploid CNV assay. (A) Schematic map of Chr4 in a diploid test strain, showing telomeres (terminal boxes), centromere (circle), and Ty retrotransposon repeats (white arrows). One homolog has an insertion of the CNV reporter near the right end, the other has TRP1 at the allelic position. (B) Reporter dosage-dependent phenotype. Cultures were serially diluted and spotted on plates. The parent strain is unable to form colonies on selective media (Copper [Cu] and Formaldehyde [FA]), while an FCR clone displays full viability. (C) Pulse field karyotypes showing a ∼1.3 Mb chromosomal rearrangement in CNV clone FCR112 (double-end arrow). The bands corresponding to normal Chr4 and Chr7 are indicated. (D) array-CGH data for FCR112. The plots show a 3x copy number amplification region on the right arm of Chr4, and a 1x deletion on the right arm of Chr7. The copy number breakpoints in both chromosomes correspond to the positions of Ty dispersed repeats. (E) Karyotype prediction for FCR112, showing a Chr7/Chr4 non-reciprocal translocation with a breakpoint junction at Ty sequences, suggesting a non-allelic homologous recombination mechanism of formation. Data credit to Ane Zeidler; full results to be described elsewhere (Zeidler, Argueso, et al., in preparation).