Back to article: Yeast-based assays for the functional characterization of cancer-associated variants of human DNA repair genes
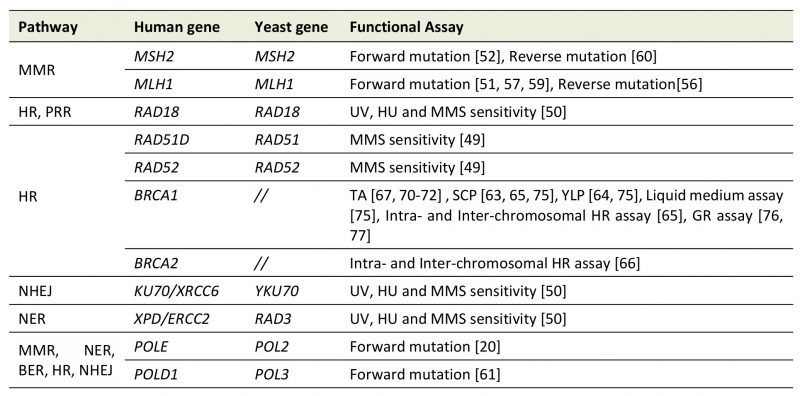
TABLE 1: DNA repair genes assayed in Saccharomyces cerevisiae.
Several yeast-based assays have been developed to characterize human genes involved in Mismatch Repair (MMR), Homologous Recombination (HR), Post Replication Repair (PRR), Base and Nucleotide Excision Repair (BER, NER), and Non-Homologous End Joining (NHEJ). The name of the yeast and human gene is shown; BRCA1 and BRCA2 have no yeast homologue. TA, transcription activation assay; SCP, Small Colony Phenotype; YLP, Yeast Localization Phenotype; GR, gene reversion; MMS, methyl methane-sulfonate, HU hydroxyurea.
20. Barbari SR, Kane DP, Moore EA, Shcherbakova PV (2018). Functional Analysis of Cancer-Associated DNA Polymerase epsilon Variants in Saccharomyces cerevisiae. G3 8(3): 1019-1029. 10.1534/g3.118.200042
41. Andersen SD, Liberti SE, Lutzen A, Drost M, Bernstein I, Nilbert M, Dominguez M, Nystrom M, Hansen TV, Christoffersen JW, Jager AC, de Wind N, Nielsen FC, Torring PM, Rasmussen LJ (2012). Functional characterization of MLH1 missense variants identified in Lynch syndrome patients. Hum Mutat 33(12): 1647-1655. 10.1002/humu.22153
50.
Kim C, Yang J, Jeong SH, Kim H, Park GH, Shin HB, Ro M, Kim KY, Park Y, Kim KP, Kwack K (2018). Yeast-based assays for characterization of the functional effects of single nucleotide polymorphisms in human DNA repair genes. PLoS One 13(3): e0193823. 10.1371/journal.pone.0193823
51. Shimodaira H, Filosi N, Shibata H, Suzuki T, Radice P, Kanamaru R, Friend SH, Kolodner RD, Ishioka C (1998). Functional analysis of human MLH1 mutations in Saccharomyces cerevisiae. Nat Genet 19(4): 384-389. 10.1038/1277
52. Gammie AE, Erdeniz N, Beaver J, Devlin B, Nanji A, Rose MD (2007). Functional characterization of pathogenic human MSH2 missense mutations in Saccharomyces cerevisiae. Genetics 177(2): 707-721. 10.1534/genetics.107.071084
56. Takahashi M, Shimodaira H, Andreutti-Zaugg C, Iggo R, Kolodner RD, Ishioka C (2007). Functional analysis of human MLH1 variants using yeast and in vitro mismatch repair assays. Cancer Res 67(10): 4595-4604. 10.1158/0008-5472.CAN-06-3509
57. Shcherbakova PV, Kunkel TA (1999). Mutator phenotypes conferred by MLH1 overexpression and by heterozygosity for mlh1 mutations. Mol Cell Biol 19(4): 3177-3183. 10.1128/mcb.19.4.3177
59. Vogelsang M, Comino A, Zupanec N, Hudler P, Komel R (2009). Assessing pathogenicity of MLH1 variants by co-expression of human MLH1 and PMS2 genes in yeast. BMC Cancer 9: 382. 10.1186/1471-2407-9-382
60. Drotschmann K, Clark AB, Tran HT, Resnick MA, Gordenin DA, Kunkel TA (1999). Mutator phenotypes of yeast strains heterozygous for mutations in the MSH2 gene. Proc Natl Acad Sci U S A 96(6): 2970-2975. 10.1073/pnas.96.6.2970
61. Daee DL, Mertz TM, Shcherbakova PV (2010). A cancer-associated DNA polymerase delta variant modeled in yeast causes a catastrophic increase in genomic instability. Proc Natl Acad Sci U S A 107(1): 157-162. 10.1073/pnas.0907526106
63. Coyne RS, McDonald HB, Edgemon K, Brody LC (2004). Functional characterization of BRCA1 sequence variants using a yeast small colony phenotype assay. Cancer Biol Ther 3(5): 453-457. 10.4161/cbt.3.5.809
64. Millot GA, Berger A, Lejour V, Boule JB, Bobo C, Cullin C, Lopes J, Stoppa-Lyonnet D, Nicolas A (2011). Assessment of human Nter and Cter BRCA1 mutations using growth and localization assays in yeast. Hum Mutat 32(12): 1470-1480. 10.1002/humu.21608
65. Caligo MA, Bonatti F, Guidugli L, Aretini P, Galli A (2009). A yeast recombination assay to characterize human BRCA1 missense variants of unknown pathological significance. Hum Mutat 30(1): 123-133. 10.1002/humu.20817
66. Spugnesi L, Balia C, Collavoli A, Falaschi E, Quercioli V, Caligo MA, Galli A (2013). Effect of the expression of BRCA2 on spontaneous homologous recombination and DNA damage-induced nuclear foci in Saccharomyces cerevisiae. Mutagenesis 28(2): 187-195. 10.1093/mutage/ges069
67. Monteiro AN, August A, Hanafusa H (1996). Evidence for a transcriptional activation function of BRCA1 C-terminal region. Proc Natl Acad Sci U S A 93(24): 13595-13599. 10.1073/pnas.93.24.13595
70. Hayes F, Cayanan C, Barilla D, Monteiro AN (2000). Functional assay for BRCA1: mutagenesis of the COOH-terminal region reveals critical residues for transcription activation. Cancer Res 60(9): 2411-2418. 10811118
71. Carvalho MA, Marsillac SM, Karchin R, Manoukian S, Grist S, Swaby RF, Urmenyi TP, Rondinelli E, Silva R, Gayol L, Baumbach L, Sutphen R, Pickard-Brzosowicz JL, Nathanson KL, Sali A, Goldgar D, Couch FJ, Radice P, Monteiro AN (2007). Determination of cancer risk associated with germ line BRCA1 missense variants by functional analysis. Cancer Res 67(4): 1494-1501. 10.1158/0008-5472.CAN-06-3297
72. Fernandes VC, Golubeva VA, Di Pietro G, Shields C, Amankwah K, Nepomuceno TC, de Gregoriis G, Abreu RBV, Harro C, Gomes TT, Silva RF, Suarez-Kurtz G, Couch FJ, Iversen ES, Monteiro ANA, Carvalho MA (2019). Impact of amino acid substitutions at secondary structures in the BRCT domains of the tumor suppressor BRCA1: Implications for clinical annotation. J Biol Chem 294(15): 5980-5992. 10.1074/jbc.RA118.005274
75. Thouvenot P, Ben Yamin B, Fourriere L, Lescure A, Boudier T, Del Nery E, Chauchereau A, Goldgar DE, Houdayer C, Stoppa-Lyonnet D, Nicolas A, Millot GA (2016). Functional Assessment of Genetic Variants with Outcomes Adapted to Clinical Decision-Making. PLoS Genet 12(6): e1006096. 10.1371/journal.pgen.1006096
76. Maresca L, Lodovichi S, Lorenzoni A, Cervelli T, Monaco R, Spugnesi L, Tancredi M, Falaschi E, Zavaglia K, Landucci E, Roncella M, Congregati C, Gadducci A, Naccarato AG, Caligo MA, Galli A (2018). Functional Interaction Between BRCA1 and DNA Repair in Yeast May Uncover a Role of RAD50, RAD51, MRE11A, and MSH6 Somatic Variants in Cancer Development. Front Genet 9: 397. 10.3389/fgene.2018.00397
77. Lodovichi S, Belle F, Cervelli T, Lorenzoni A, Maresca L, Cozzani C, Caligo MA, Galli A (2020). Effect of BRCA1 missense variants on gene reversion in DNA double-strand break repair mutants and cell cycle-arrested cells of Saccharomyces cerevisiae. Mutagenesis 35(2): 189-195. 10.1093/mutage/gez043