Back to article: Variants of the human RAD52 gene confer defects in ionizing radiation resistance and homologous recombination repair in budding yeast
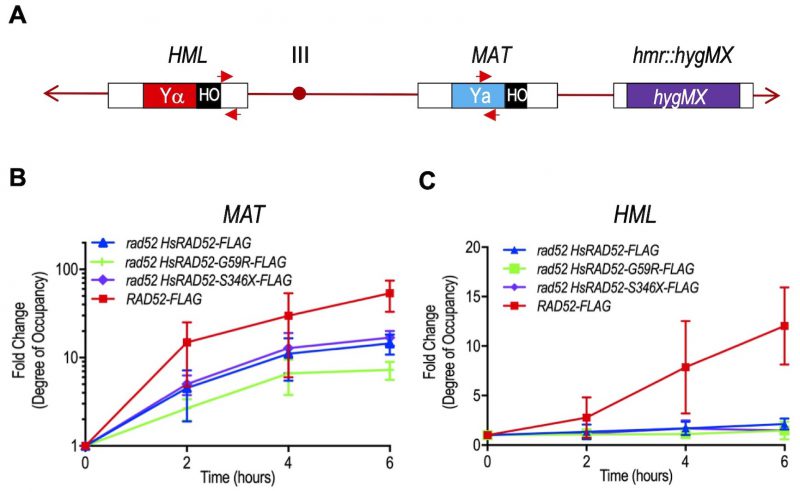
FIGURE 6: The adh1::HsRAD52-G59R-FLAG and adh1::HsRAD52-S346X-FLAG alleles have differential effects on the interaction of HsRAD52 with the MAT and HML loci during MTI. (A) Cartoon depicting substrates for MTI and location of primers used for quantitation of immunoprecipitated genomic sequences. Following DSB formation by HO endonuclease at the HO cut site (black box) at the MAT locus on chromosome III, exonucleolytic processing results in the accumulation of ssDNA in flanking sequences. ScRad52-FLAG, or HsRAD52-FLAG progressively associate with the ssDNA. This association facilitates retention of the DNA sequences by ChIP, which are quantitated by qPCR using MATa recipient primers (red arrows; Table S3) complementary to a 110 bp sequence laying 500 bp upstream from the DSB at MAT. Deletion of the HMR locus and replacement with a hygMX marker (purple box) removes additional complementary sequences from the genome. After association with ssDNA at MAT, the HRR apparatus facilitates a search for homologous genomic sequences that gives rise to heteroduplex formation with the intact HML locus. The sequences that lay proximal to the HO cut site at the border of Yα (red box) are the putative initial location for heteroduplex formation. Association of ScRad52-FLAG or HsRAD52-FLAG with the heteroduplex results in retention of these sequences by ChIP, and their quantitation is done by qPCR using HMLα donor primers (red arrows; Table S3) complementary to a 187 bp sequence laying 67 bp downstream from the HO cut site sequence at HML. (B) ScRad52-FLAG and HsRAD52-FLAG display similar kinetics of association with sequences at the MAT locus after DSB formation. Single colonies of wild-type (ABX3961-4C), rad52 (ABX3943-3B), rad52 adh1::HsRAD52-FLAG (ABX3977-10C), rad52 adh1::HsRAD52-G59R-FLAG (ABX3985-55B), and rad52 adh1::HsRAD52-S346X-FLAG (ABX3994-18D) strains carrying MTI assay components were used to establish cultures from which aliquots were collected at various times before and after DSB formation at the MAT locus by HO endonuclease. Whole cell extracts were prepared, subjected to ChIP using anti-FLAG antibody and the immunoprecipitated DNA from the MAT (experimental) and SAM1 (control) loci quantitated by qPCR. Fold changes in degree of occupancy of the FLAG-tagged proteins relative to those observed before DSB formation were normalized to a control strain lacking FLAG-tagged proteins (ABX3933-46C). Mean fold changes from at least eight determinations using DNA collected from at least three independent time courses, and standard deviations were plotted against elapsed time after initiation of DSB formation. (C) HsRAD52-G59R-FLAG and HsRAD52-S346X-FLAG display defects in association with the HMR locus during MTI. Same as above except immunoprecipitated DNA from the HMR locus (experimental) was quantitated by qPCR.