Back to article: Microfluidic techniques for separation of bacterial cells via taxis
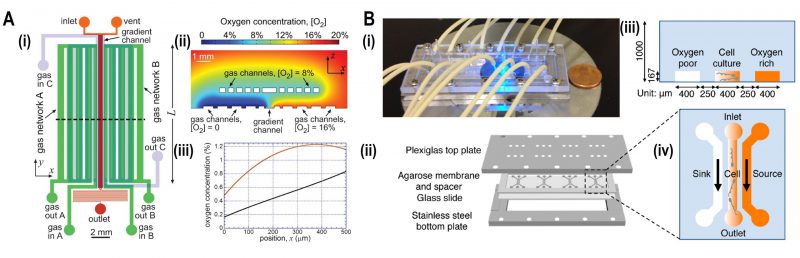
FIGURE 5: Microfluidic devices for bacterial aerotaxis. (A) Two-layered PDMS chip for generating oxygen gradients. (i) Lower layer supplied with gases by an in-plane gas channel network (A and B) and the top layer with gases by an out of plane gas channel network. (ii) Oxygen-diffusion simulation demonstrated that the oxygen gradient was generated ranging from 0 – 20% inside the chip. (iii) Simulation plot of oxygen concentration versus position 'x,' i.e., cross-section view of the gradient inside the channel. Incorporation of gas channel C in the device generates near-linear stable oxygen gradient (black line) in comparison with the condition of no gas channel C (Red curve). Reproduced from [44]. (B) Agarose-based chemical or oxygen gradient generator. (i) A microfluidic device on the microscopic stage with tubing to inject media, containing oxidized flavin (chemo effector) or oxygen. (ii) Four three-channel microstructures were engraved in agarose membrane which was clamped by plexiglass plate at the top and stainless-steel plate at the bottom which was spaced by a glass slide. (iii) Schematic diagram of a single three-channel microstructure, consisting of the middle channel for the bacterial culture which was flanked by side channels for either oxygen or oxidized flavin to generate a gradient across the middle channel. (iv) Side view of the channels with 250 μm spacing where oxygen-rich channel (coloured) and oxygen-poor channel (colourless) were juxtaposed to the middle channel containing an oxygen gradient. Reproduced from [46].
44. Adler M, Erickstad M, Gutierrez E, and Groisman A (2012). Studies of bacterial aerotaxis in a microfluidic device. Lab Chip 12(22): 4835–4847. doi: 10.1039/C2LC21006A
46. Kim BJ, Chu I, Jusuf S, Kuo T, TerAvest MA, Angenent LT, and Wu M (2016). Oxygen Tension and Riboflavin Gradients Cooperatively Regulate the Migration of Shewanella oneidensis MR-1 Revealed by a Hydrogel-Based Microfluidic Device. Front Microbiol 7. doi: 10.3389/fmicb.2016.01438