Back to article: Regulation of Cdc42 for polarized growth in budding yeast
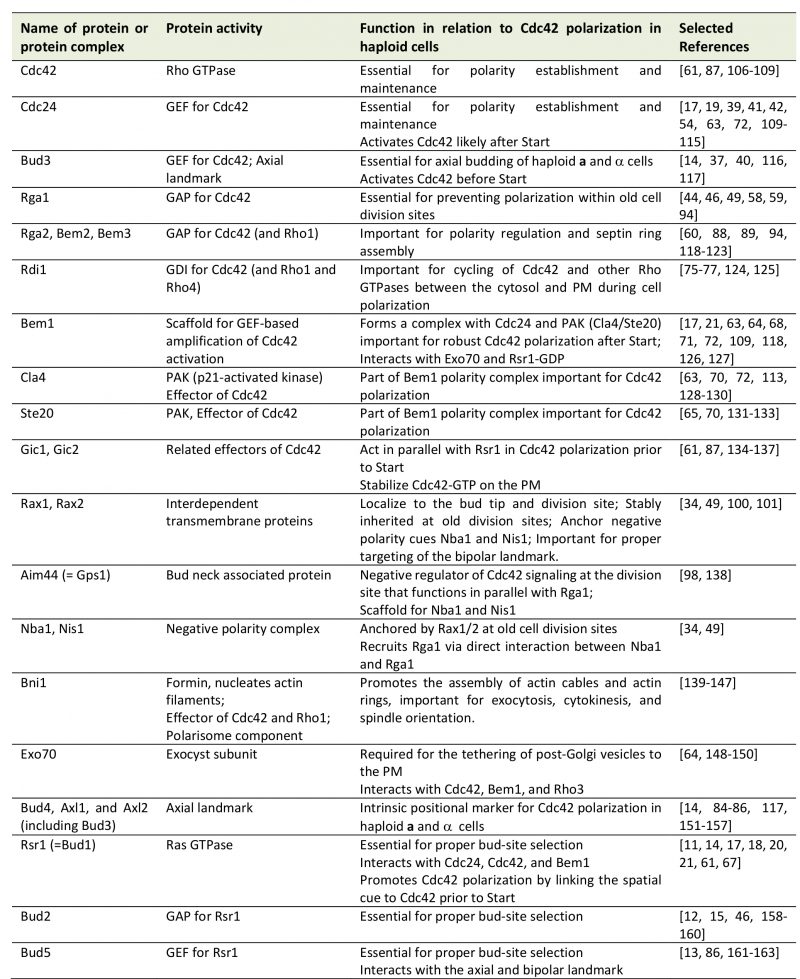
TABLE 1. The Cdc42 and Rsr1 GTPses and their regulators and effectors.
11. Bender A, Pringle JR (1989). Multicopy suppression of the cdc24 budding defect in yeast by CDC42 and three newly identified genes including the ras-related gene RSR1. Proc Natl Acad Sci USA 86(24): 9976-9980. 10.1073/pnas.86.24.9976
14. Chant J, Herskowitz I (1991). Genetic control of bud site selection in yeast by a set of gene products that constitute a morphogenetic pathway. Cell 65(7): 1203-1212. 10.1016/0092-8674(91)90015-q
17. Park H-O, Bi E, Pringle JR, Herskowitz I (1997). Two active states of the Ras-related Bud1/Rsr1 protein bind to different effectors to determine yeast cell polarity. Proc Natl Acad Sci USA 94(9): 4463-4468. 10.1073/pnas.94.9.4463
18. Kozminski KG, Beven L, Angerman E, Tong AHY, Boone C, Park H-O (2003). Interaction between a Ras and a Rho GTPase couples selection of a growth site to the development of cell polarity in yeast. Mol Biol Cell 14(12): 4958-4970. 10.1091/mbc.e03-06-0426
19. Shimada Y, Wiget P, Gulli MP, Bi E, Peter M (2004). The nucleotide exchange factor Cdc24p may be regulated by auto-inhibition. EMBO J 23(5): 1051-1062. 10.1038/sj.emboj.7600124
20. Kang PJ, Beven L, Hariharan S, Park H-O (2010). The Rsr1/Bud1 GTPase Interacts with Itself and the Cdc42 GTPase during Bud-Site Selection and Polarity Establishment in Budding Yeast. Mol Biol Cell 21(17): 3007-3016. 10.1091/mbc.E10-03-0232
21. Miller KE, Lo WC, Chou CS, Park HO (2019). Temporal regulation of cell polarity via the interaction of the Ras GTPase Rsr1 and the scaffold protein Bem1. Mol Biol Cell 30(20): 2543-2557. 10.1091/mbc.E19-02-0106
34. Meitinger F, Khmelinskii A, Morlot S, Kurtulmus B, Palani S, Andres-Pons A, Hub B, Knop M, Charvin G, Pereira G (2014). A memory system of negative polarity cues prevents replicative aging. Cell 159(5): 1056-1069. 10.1016/j.cell.2014.10.014
37. Kang PJ, Lee ME, Park H-O (2014). Bud3 activates Cdc42 to establish a proper growth site in budding yeast. J Cell Biol 206(1): 19-28. 10.1083/jcb.201402040
39. Zheng Y, Cerione R, Bender A (1994). Control of the yeast bud-site assembly GTPase Cdc42: catalysis of guanine nucleotide exchange by Cdc24 and stimulation of GTPase activity by Bem3. J Biol Chem 269(4): 2369-2372. 8300560
40. Chant J, Mischke M, Mitchell E, Herskowitz I, Pringle JR (1995). Role of Bud3p in producing the axial budding pattern of yeast. J Cell Biol 129(3): 767-778. 10.1083/jcb.129.3.767
41. Nern A, Arkowitz RA (2000). Nucleocytoplasmic shuttling of the Cdc42p exchange factor Cdc24p. J Cell Biol 148(6): 1115-1122. 10.1083/jcb.148.6.1115
42. Shimada Y, Gulli M-P, Peter M (2000). Nuclear sequestration of the exchange factor Cdc24p by Far1 regulates cell polarity during mating. Nat Cell Biol 2(2): 117-124. 10.1038/35000073
44. Tong Z, Gao XD, Howell AS, Bose I, Lew DJ, Bi E (2007). Adjacent positioning of cellular structures enabled by a Cdc42 GTPase-activating protein-mediated zone of inhibition. J Cell Biol 179(7): 1375-1384. 10.1083/jcb.200705160
46. Lee ME, Lo WC, Miller KE, Chou CS, Park H-O (2015). Regulation of Cdc42 polarization by the Rsr1 GTPase and Rga1, a Cdc42 GTPase-activating protein, in budding yeast. J Cell Sci 128(11): 2106-2117. 10.1242/jcs.166538
49. Miller KE, Lo WC, Lee ME, Kang PJ, Park H-O (2017). Fine-tuning the orientation of the polarity axis by Rga1, a Cdc42 GTPase-activating protein. Mol Biol Cell 28(26): 3773-3788. 10.1091/mbc.E17-01-0074
54. Toenjes KA, Sawyer MM, Johnson DI (1999). The guanine-nucleotide-exchange factor Cdc24p is targeted to the nucleus and polarized growth sites. Curr Biol 9(20): 1183-1186. 10.1016/S0960-9822(00)80022-6
58. Stevenson BJ, Ferguson B, De Virgilio C, Bi E, Pringle JR, Ammerer G, Sprague GFJ (1995). Mutation of RGA1, which encodes a putative GTPase-activating protein for the polarity-establishment protein Cdc42p, activates the pheromone-response pathway in the yeast Saccharomyces cerevisiae. Genes Dev 9(23): 2949-2963. 10.1101/gad.9.23.2949
59. Chen GC, Zheng L, Chan CS (1996). The LIM domain-containing Dbm1 GTPase-activating protein is required for normal cellular morphogenesis in Saccharomyces cerevisiae. Mol Cell Biol 16(4): 1376-1390. 10.1128/mcb.16.4.1376
60. Smith GR, Givan SA, Cullen P, Sprague GF, Jr. (2002). GTPase-activating proteins for Cdc42. Eukaryot Cell 1(3): 469-480. 10.1128/ec.1.3.469-480.2002
61. Kang PJ, Miller KE, Guegueniat J, Beven L, Park H-O (2018). The shared role of the Rsr1 GTPase and Gic1/Gic2 in Cdc42 polarization. Mol Biol Cell 29(20): 2359-2369. 10.1091/mbc.E18-02-0145
63. Witte K, Strickland D, Glotzer M (2017). Cell cycle entry triggers a switch between two modes of Cdc42 activation during yeast polarization. eLife 6: e26722. 10.7554/eLife.26722
64. Liu D, Novick P (2014). Bem1p contributes to secretory pathway polarization through a direct interaction with Exo70p. J Cell Biol 207(1): 59-72. 10.1083/jcb.201404122
65. Moran KD, Kang H, Araujo AV, Zyla TR, Saito K, Tsygankov D, Lew DJ (2019). Cell-cycle control of cell polarity in yeast. J Cell Biol 218(1): 171-189. 10.1083/jcb.201806196
67. Park H-O, Kang PJ, Rachfal AW (2002). Localization of the Rsr1/Bud1 GTPase involved in selection of a proper growth site in yeast. J Biol Chem 277(30): 26721-26724. 10.1074/jbc.C200245200
68. Irazoqui JE, Gladfelter AS, Lew DJ (2003). Scaffold-mediated symmetry breaking by Cdc42p. Nat Cell Biol 5(12): 1062-1070. 10.1038/ncb1068
70. Kozubowski L, Saito K, Johnson JM, Howell AS, Zyla TR, Lew DJ (2008). Symmetry-breaking polarization driven by a Cdc42p GEF-PAK complex. Curr Biol 18(22): 1719-1726. 10.1016/j.cub.2008.09.060
71. Smith SE, Rubinstein B, Mendes Pinto I, Slaughter BD, Unruh JR, Li R (2013). Independence of symmetry breaking on Bem1-mediated autocatalytic activation of Cdc42. J Cell Biol 202(7): 1091-1106. 10.1083/jcb.201304180
72. Rapali P, Mitteau R, Braun C, Massoni-Laporte A, Unlu C, Bataille L, Arramon FS, Gygi SP, McCusker D (2017). Scaffold-mediated gating of Cdc42 signalling flux. eLife 6: e25257. 10.7554/eLife.25257
75. Slaughter BD, Das A, Schwartz JW, Rubinstein B, Li R (2009). Dual modes of cdc42 recycling fine-tune polarized morphogenesis. Dev Cell 17(6): 823-835. 10.1016/j.devcel.2009.10.022
76. Freisinger T, Klunder B, Johnson J, Muller N, Pichler G, Beck G, Costanzo M, Boone C, Cerione RA, Frey E, Wedlich-Soldner R (2013). Establishment of a robust single axis of cell polarity by coupling multiple positive feedback loops. Nat Commun 4(1807): 1807. 10.1038/ncomms2795
77. Woods B, Lai H, Wu CF, Zyla TR, Savage NS, Lew DJ (2016). Parallel Actin-Independent Recycling Pathways Polarize Cdc42 in Budding Yeast. Curr Biol 26(16): 2114-2126. 10.1016/j.cub.2016.06.047
84. Sanders SL, Herskowitz I (1996). The Bud4 protein of yeast, required for axial budding, is localized to the mother/bud neck in a cell cycle-dependent manner. J Cell Biol 134(2): 413-427. 10.1083/jcb.134.2.413
85. Kang PJ, Angerman E, Jung CH, Park H-O (2012). Bud4 mediates the cell-type-specific assembly of the axial landmark in budding yeast. J Cell Sci 125(Pt 16): 3840-3849. 10.1242/jcs.103697
86. Kang PJ, Sanson A, Lee B, Park H-O (2001). A GDP/GTP exchange factor involved in linking a spatial landmark to cell polarity. Science 292(5520): 1376-1378. 10.1126/science.1060360
87. Iwase M, Luo J, Nagaraj S, Longtine M, Kim HB, Haarer BK, Caruso C, Tong Z, Pringle JR, Bi E (2006). Role of a Cdc42p effector pathway in recruitment of the yeast septins to the presumptive bud site. Mol Biol Cell 17(3): 1110-1125. 10.1091/mbc.e05-08-0793
88. Gladfelter AS, Bose I, Zyla TR, Bardes ESG, Lew DJ (2002). Septin ring assembly involves cycles of GTP loading and hydrolysis by Cdc42p. J Cell Biol 156(2): 315-326. 10.1083/jcb.200109062
89. Okada S, Leda M, Hanna J, Savage NS, Bi E, Goryachev AB (2013). Daughter cell identity emerges from the interplay of Cdc42, septins, and exocytosis. Dev Cell 26(2): 148-161. 10.1016/j.devcel.2013.06.015
94. Caviston JP, Longtine M, Pringle JR, Bi E (2003). The role of Cdc42p GTPase-activating proteins in assembly of the septin ring in yeast. Mol Biol Cell 14(10): 4051-4066. 10.1091/mbc.e03-04-0247
98. Meitinger F, Richter H, Heisel S, Hub B, Seufert W, Pereira G (2013). A safeguard mechanism regulates Rho GTPases to coordinate cytokinesis with the establishment of cell polarity. PLoS Biol 11(2): e1001495. 10.1371/journal.pbio.1001495
100. Chen T, Hiroko T, Chaudhuri A, Inose F, Lord M, Tanaka S, Chant J, Fujita A (2000). Multigenerational cortical inheritance of the Rax2 protein in orienting polarity and division in yeast. Science 290(5498): 1975-1978. 10.1126/science.290.5498.1975
101. Kang PJ, Angerman E, Nakashima K, Pringle JR, Park H-O (2004). Interactions among Rax1p, Rax2p, Bud8p, and Bud9p in marking cortical sites for bipolar bud-site selection in yeast. Mol Biol Cell 15(11): 5145-5157. 10.1091/mbc.e04-07-0600
106. Adams AE, Johnson DI, Longnecker RM, Sloat BF, Pringle JR (1990). CDC42 and CDC43, two additional genes involved in budding and the establishment of cell polarity in the yeast Saccharomyces cerevisiae. J Cell Biol 111(1): 131-142. 10.1083/jcb.111.1.131
107. Johnson DI, Pringle JR (1990). Molecular characterization of CDC42, a Saccharomyces cerevisiae gene involved in the development of cell polarity. J Cell Biol 111(1): 143-152. 10.1083/jcb.111.1.143
108. Sartorel E, Unlu C, Jose M, Massoni-Laporte A, Meca J, Sibarita JB, McCusker D (2018). Phosphatidylserine and GTPase activation control Cdc42 nanoclustering to counter dissipative diffusion. Mol Biol Cell 29(11): 1299-1310. 10.1091/mbc.E18-01-0051
109. Meca J, Massoni-Laporte A, Martinez D, Sartorel E, Loquet A, Habenstein B, McCusker D (2019). Avidity-driven polarity establishment via multivalent lipid-GTPase module interactions. EMBO J 38(3): e99652. 10.15252/embj.201899652
110. Sloat B, Adams A, Pringle JR (1981). Roles of the CDC24 gene product in cellular morphogenesis during the Saccharomyces cerevisiae cell cycle. J Cell Biol 89(3):395-405. 10.1083/jcb.89.3.395
111. Nern A, Arkowitz RA (1998). A GTP-exchange factor required for cell orientation. Nature 391(6663):195-198. 10.1038/34458
112. Nern A, Arkowitz RA (1999). A Cdc24p-Far1p-Gbetagamma protein complex required for yeast orientation during mating. J Cell Biol 144(6):1187-1202. 10.1083/jcb.144.6.1187
113. Gulli MP, Jaquenoud M, Shimada Y, Niederhauser G, Wiget P, Peter M (2000). Phosphorylation of the Cdc42 exchange factor Cdc24 by the PAK-like kinase Cla4 may regulate polarized growth in yeast. Mol Cell 6(5):1155-1167. 10.1016/s1097-2765(00)00113-1
114. Wiget P, Shimada Y, Butty AC, Bi E, Peter M (2004). Site-specific regulation of the GEF Cdc24p by the scaffold protein Far1p during yeast mating. EMBO J 23(5):1063-1074. 10.1038/sj.emboj.7600123
115. Mionnet C, Bogliolo S, Arkowitz RA (2008). Oligomerization regulates the localization of Cdc24, the Cdc42 activator in Saccharomyces cerevisiae. J Biol Chem 283(25): 17515-17530. 10.1074/jbc.M800305200
116. Lord M, Yang MC, Mischke M, Chant J (2000). Cell cycle programs of gene expression control morphogenetic protein localization. J Cell Biol 151(7):1501-1512. 10.1083/jcb.151.7.1501
117. Chen X, Wang K, Svitkina T, Bi E (2020). Critical Roles of a RhoGEF-Anillin Module in Septin Architectural Remodeling during Cytokinesis. Curr Biol 30(8): 1477-1490.e1473. 10.1016/j.cub.2020.02.023
118. Bender A, Pringle JR (1991). Use of a screen for synthetic lethal and multicopy suppressee mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. Mol Cell Biol 11(3): 1295-1305. 10.1128/mcb.11.3.1295
119. Kim Y-J, Francisco L, Chen G-C, Marcotte E, Chan CSM (1994). Control of cellular morphogenesis by the Ipl2/Bem2 GTPase-activating protein: Possible role of protein phosphorylation. J Cell Biol 127(5):1381-1394. 10.1083/jcb.127.5.1381
120. Peterson J, Zheng Y, Bender L, Myers A, Cerione R, Bender A (1994). Interactions between the bud emergence proteins Bem1p and Bem2p and Rho-type GTPases in yeast. J Cell Biol 127(5):1395-1406. 10.1083/jcb.127.5.1395
121. Knaus M, Pelli-Gulli M-P, van Drogen F, Springer S, Jaquenoud M, Peter M (2007). Phosphorylation of Bem2p and Bem3p may contribute to local activation of Cdc42p at bud emergence. EMBO J 26(21): 4501-4513. 10.1038/sj.emboj.7601873
122. Sopko R, Huang D, Smith JC, Figeys D, Andrews BJ (2007). Activation of the Cdc42p GTPase by cyclin-dependent protein kinases in budding yeast. EMBO J 26(21): 4487-4500. 10.1038/sj.emboj.7601847
123. Mukherjee D, Sen A, Boettner DR, Fairn GD, Schlam D, Bonilla Valentin FJ, Michael McCaffery J, Hazbun T, Staiger CJ, Grinstein S, Lemmon SK, Claudio Aguilar R (2013). Bem3, a Cdc42 GTPase-activating protein, traffics to an intracellular compartment and recruits the secretory Rab GTPase Sec4 to endomembranes. J Cell Sci 126(Pt 20): 4560-4571. 10.1242/jcs.117663
124. Masuda T, Tanaka K, Nonaka H, Yamochi W, Maeda A, Takai Y (1994). Molecular cloning and characterization of yeast rho GDP dissociation inhibitor. J Biol Chem 269(31):19713-19718. 8051050
125. Koch G, Tanaka K, Masuda T, Yamochi W, Nonaka H, Takai Y (1997). Association of the Rho family small GTP-binding proteins with Rho GDP dissociation inhibitor (Rho GDI) in Saccharomyces cerevisiae. Oncogene 15(4):417-422. 10.1038/sj.onc.1201194
126. Chenevert J, Corrado K, Bender A, Pringle J, Herskowitz I (1992). A yeast gene (BEM1) necessary for cell polarization whose product contains two SH3 domains. Nature 356(6364):77-79. 10.1038/356077a0
127. Bose I, Irazoqui JE, Moskow JJ, Bardes ES, Zyla TR, Lew DJ (2001). Assembly of scaffold-mediated complexes containing Cdc42p, the exchange factor Cdc24p, and the effector Cla4p required for cell cycle-regulated phosphorylation of Cdc24p. J Biol Chem 276(10): 7176-7186. 10.1074/jbc.M010546200
128. Cvrckova F, De Virgilio C, Manser E, Pringle JR, Nasmyth K (1995). Ste20-like protein kinases are required for normal localization of cell growth and for cytokinesis in budding yeast. Genes Dev 9(15):1817-1830. 10.1101/gad.9.15.1817
129. Benton BK, Tinkelenberg A, Gonzalez I, Cross FR (1997). Cla4p, a Saccharomyces cerevisiae Cdc42p-activated kinase involved in cytokinesis, is activated at mitosis. Mol Cell Biol 17(9):5067-5076. 10.1128/mcb.17.9.5067
130. Weiss EL, Bishop AC, Shokat KM, Drubin DG (2000). Chemical genetic analysis of the budding-yeast p21-activated kinase Cla4p. Nat Cell Biol 2(10):677 – 685. 10.1038/35036300
131. Leberer E, Dignard D, Harcus D, Thomas DY, Whiteway M (1992). The protein kinase homologue Ste20p is required to link the yeast pheromone response G-protein beta gamma subunits to downstream signalling components. EMBO J 11(13): 4815-4824. 10.1002/j.1460-2075.1992.tb05587.x
132. Peter M, Neiman A, Park H-O, Lohuizen Mv, Herskowitz I (1996). Interaction between the small GTP-binding protein CDC42 and the protein kinase STE20 is necessary for signal transduction in the yeast pheromone pathway. EMBO J 15(24):7046-7059. 9003780
133. Atkins BD, Yoshida S, Saito K, Wu CF, Lew DJ, Pellman D (2013). Inhibition of Cdc42 during mitotic exit is required for cytokinesis. J Cell Biol 202(2): 231-240. 10.1083/jcb.201301090
134. Chen GC, Kim YJ, Chan CS (1997). The Cdc42 GTPase-associated proteins Gic1 and Gic2 are required for polarized cell growth in Saccharomyces cerevisiae. Genes Dev 11: 2958-2971. 10.1101/gad.11.22.2958
135. Brown JL, Jaquenoud M, Gulli MP, Chant J, Peter M (1997). Novel Cdc42-binding proteins Gic1 and Gic2 control cell polarity in yeast. Genes Dev 11(22):2972-2982. 10.1101/gad.11.22.2972
136. Jaquenoud M, Gulli MP, Peter K, Peter M (1998). The Cdc42p effector Gic2p is targeted for ubiquitin-dependent degradation by the SCFGrr1 complex. EMBO J 17(18):5360-5373. 10.1093/emboj/17.18.5360
137. Sadian Y, Gatsogiannis C, Patasi C, Hofnagel O, Goody RS, Farkasovsky M, Raunser S (2013). The role of Cdc42 and Gic1 in the regulation of septin filament formation and dissociation. eLife 2: e01085. 10.7554/eLife.01085
138. Meitinger F, Pereira G (2017). The septin-associated kinase Gin4 recruits Gps1 to the site of cell division. Mol Biol Cell 28(7): 883-889. 10.1091/mbc.E16-09-0687
139. Kohno H, Tanaka K, Mino A, Umikawa M, Imamura H, Fujiwara T, Fujita Y, Hotta K, Qadota H, Watanabe T, Ohya Y, Takai Y (1996). Bni1p implicated in cytoskeletal control is a putative target of Rho1p small GTP binding protein in Saccharomyces cerevisiae.EMBO J 15(22):6060-6068. 10.1002/j.1460-2075.1996.tb00994.x
140. Evangelista M, Blundell K, Longtine MS, Chow CJ, Adames N, Pringle JR, Peter M, Boone C (1997). Bni1p, a yeast formin linking Cdc42p and the actin cytoskeleton during polarized morphogenesis. Science 276(5309): 118-122. 10.1126/science.276.5309.118
141. Lee L, Klee SK, Evangelista M, Boone C, Pellman D (1999). Control of mitotic spindle position by the Saccharomyces cerevisiae formin Bni1p. J Cell Biol 144(5): 947-961. 10.1083/jcb.144.5.947
142. Pruyne D, Evangelista M, Yang C, Bi E, Zigmond S, Bretscher A, Boone C (2002). Role of formins in actin assembly: nucleation and barbed-end association. Science 297(5581):612-615. 10.1126/science.1072309
143. Sagot I, Rodal AA, Moseley J, Goode BL, Pellman D (2002). An actin nucleation mechanism mediated by Bni1 and profilin. Nat Cell Biol 4(8):626-631. 10.1038/ncb834
144. Moseley JB, Goode BL (2005). Differential activities and regulation of Saccharomyces cerevisiae formin proteins Bni1 and Bnr1 by Bud6. J Biol Chem 280(30): 28023-28033. 10.1038/ncb834
145. Liu W, Santiago-Tirado FH, Bretscher A (2012). Yeast formin Bni1p has multiple localization regions that function in polarized growth and spindle orientation. Mol Biol Cell 23(3): 412-422. 10.1091/mbc.E11-07-0631
146. Miao Y, Wong CC, Mennella V, Michelot A, Agard DA, Holt LJ, Yates JR, 3rd, Drubin DG (2013). Cell-cycle regulation of formin-mediated actin cable assembly. Proc Natl Acad Sci USA 110(47): E4446-4455. 10.1073/pnas.1314000110
147. Feng Z, Okada S, Cai G, Zhou B, Bi E (2015). MyosinII heavy chain and formin mediate the targeting of myosin essential light chain to the division site before and during cytokinesis. Mol Biol Cell 26(7): 1211-1224. 10.1091/mbc.E14-09-1363
148. TerBush DR, Maurice T, Roth D, Novick P (1996). The Exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. EMBO J 15(23): 6483-6494. 10.1002/j.1460-2075.1996.tb01039.x
149. Robinson NG, Guo L, Imai J, Toh-E A, Matsui Y, Tamanoi F (1999). Rho3 of Saccharomyces cerevisiae, Which Regulates the Actin Cytoskeleton and Exocytosis, Is a GTPase Which Interacts with Myo2 and Exo70. Mol Cell Biol 19(5):3580-3587. 10.1128/mcb.19.5.3580
150. Boyd C, Hughes T, Pypaert M, Novick P (2004). Vesicles carry most exocyst subunits to exocytic sites marked by the remaining two subunits, Sec3p and Exo70p. J Cell Biol 167(5): 889-901. 10.1083/jcb.200408124
151. Fujita A, Oka C, Arikawa Y, Katagai T, Tonouchi A, Kuhara S, Misumi Y (1994). A yeast gene necessary for bud-site selection encodes a protein similar to insulin-degrading enzymes. Nature 372(6506):567-570. 10.1038/372567a0
152. Roemer T, Madden K, Chang J, Snyder M (1996). Selection of axial growth sites in yeast requires Axl2p, a novel plasma membrane glycoprotein. Genes Dev 10(7):777-793. 10.1101/gad.10.7.777
153. Halme A, Michelitch M, Mitchell EL, Chant J (1996). Bud10p directs axial cell polarization in budding yeast and resembles a transmembrane receptor. Curr Biol 6(5):570-579. 10.1016/s0960-9822(02)00543-2
154. Sanders SL, Gentzsch M, Tanner W, Herskowitz I (1999). O-glycosylation of Axl2/Bud10p by Pmt4p is required for its stability, localization, and function in daughter cells. J Cell Biol 145(6):1177-1188. 10.1083/jcb.145.6.1177
155. Lord M, Inose F, Hiroko T, Hata T, Fujita A, Chant J (2002). Subcellular localization of Axl1, the cell type-specific regulator of polarity. Curr Biol 12(15):1347-1352. 10.1016/s0960-9822(02)01042-4
156. Gao XD, Sperber LM, Kane SA, Tong Z, Tong AH, Boone C, Bi E (2007). Sequential and distinct roles of the cadherin domain-containing protein Axl2p in cell polarization in yeast cell cycle. Mol Biol Cell 18(7): 2542-2560. 10.1091/mbc.e06-09-0822
157. Kang PJ, Hood-DeGrenier JK, Park H-O (2013). Coupling of septins to the axial landmark by Bud4 in budding yeast. J Cell Sci 126:1218-1226. 10.1242/jcs.118521