Back to article: Structural insights into the architecture and assembly of eukaryotic flagella
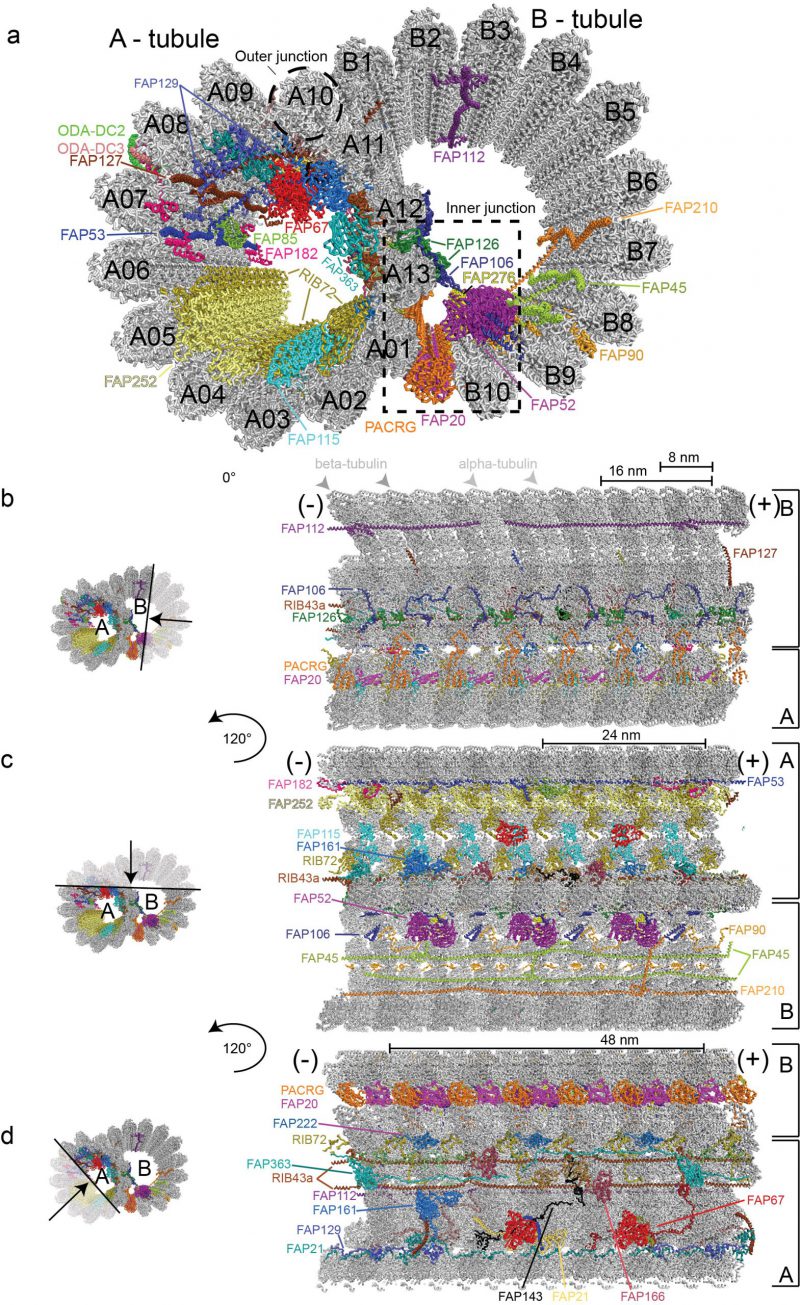
FIGURE 2: Structure of the 48nm ciliary doublet microtubule repeat (pdb entry: 6u42). (a) Cross section through a ciliary DMT. The outer dynein arms docking complexes 2 and 3 serve as docking sites for ODAs on the exterior of the DMTs and are labelled ODA-DC2 and ODA-DC3. Inner microtubule proteins (MIPs) are highlighted in different colours and labelled according to protein name. (b-d) Depiction of microtubule inner proteins, their interaction network and periodicity visualized within the confinements of the doublet microtubule from the minus (-) to the plus (+) end. (b) Lateral view of MIPs decorating the lumen of B-tubule as seen after a 10 nm deep slice facing the viewer was removed. The black arrow in the transparent region on the left representation indicates the region that has been removed for better clarity. The ODA-DC complex is left out as part of the removed section. (c) A lateral view displaying MIPs decorating both the A- and B-tubules. A 10 nm deep slice facing the viewer was removed as shown by the black arrow in the transparent region on the left representation. MIPs are visualized after rotation of the DMTs of 120° along the longitudinal axis relative to (b). (d) Visualisation of MIPs decorating the lumen of the A-tubule after removal of a 10 nm deep slice facing the viewer and rotation of the doublet microtubule with 120° along its longitudinal axis relative to (c). In this representation the alternating PCRG/FAP20 complex is observed from the exterior of the B-tubule.