Back to article: Systematic analysis of nuclear gene function in respiratory growth and expression of the mitochondrial genome in S. cerevisiae
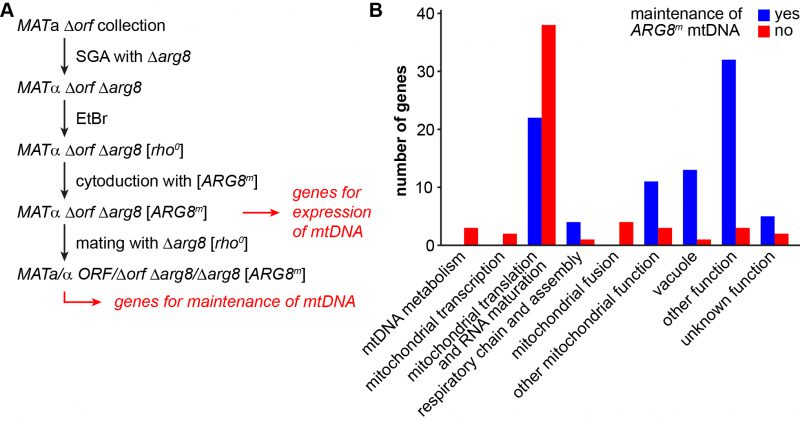
FIGURE 3: Defining the genes required for expression and maintenance of the mitochondrial genome. (A) Flow chart depicting the outline of the experiment. A Δorf Δarg8 double mutant collection was generated using SGA technology (see methods). The mitochondrial genome was eliminated from all double mutants by treatment with EtBr. A functional mtDNA containing the ARG8m allele was introduced into all strains by cytoduction. Mutants that were unable to grow on media lacking arginine after cytoduction were considered to have lost their mtDNA or to be unable to express the ARG8m gene. To test for this, the resulting Δorf Δarg8 [ARG8m] double mutant collection was crossed with a Δarg8 [rho0] strain and the resulting diploid strains were scored for growth on media lacking arginine. Mutants that were unable to grow were considered to suffer from mtDNA instability. See text for details. (B) The mutants that had lost their mtDNA or that were unable to express the ARG8m gene were manually grouped into functional categories. Depicted is how often each functional group is represented among these two sets of mutants. Blue bars represent mutants that maintained the [ARG8m] mitochondrial genome, but were unable to express Arg8m (i.e. the genes listed in Table 3; these are the “genes for expression of mtDNA” minus “genes for maintenance of mtDNA” in panel A). Red bars represent mutants that lost the [ARG8m] mitochondrial genome (i.e. the genes listed in Table 4; these are the “genes for maintenance of mtDNA” in panel A that could be confirmed by DAPI staining).