Back to article: Urm1, not quite a ubiquitin-like modifier?
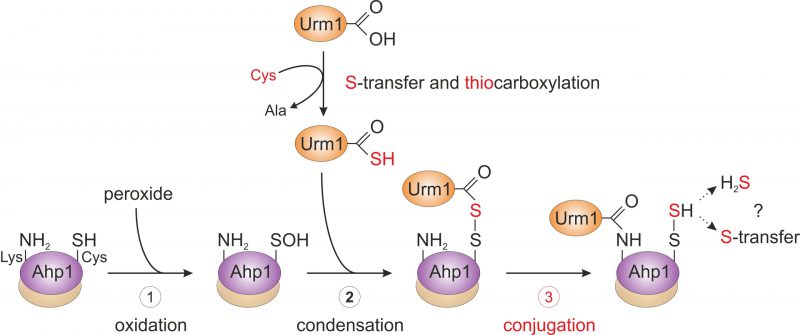
FIGURE 3: Working model for urmylation of the peroxiredoxin Ahp1. Step 1 (oxidation): the thiol of the peroxidatic cysteine (Ahp1-SH) is oxidized by the reaction with a peroxide to form the sulfenic acid (Ahp1-SOH) that becomes surface exposed and accessible for the reaction with Urm1. Step 2 (condensation): sulfur mobilization from cysteine and S-transfer to Urm1 yields the activated Urm1 thiocarboxylate (Urm1-COSH). This condenses with the sulfenic acid to form an acyl disulfide (Ahp1-S-S-Urm1). Step 3 (conjugation): the ε-amino group of a nearby lysine residue in Ahp1 (Ahp1-NH2) mounts a nucleophilic attack on the Urm1 carbonyl group, generating an iso-peptide bond with Urm1 (Ahp1-NH-CO-Urm1) and a persulfide on the peroxidatic cysteine (Ahp1-S-SH). Possibly, Ahp1 transfers the persulfide to other proteins or releases hydrogen sulfide (H2S). The lysine-directed urmylation might support the S-transfer by sterically preventing the latter option. For simplicity, the illustration solely involves one subunit of the Ahp1 homodimer. Partial reaction steps in need of further verification are marked in red (step 3), while those supported by experimental evidence (step 1 and 2) are labeled black.