Back to article: Genome, transcriptome and secretome analyses of the antagonistic, yeast-like fungus Aureobasidium pullulans to identify potential biocontrol genes
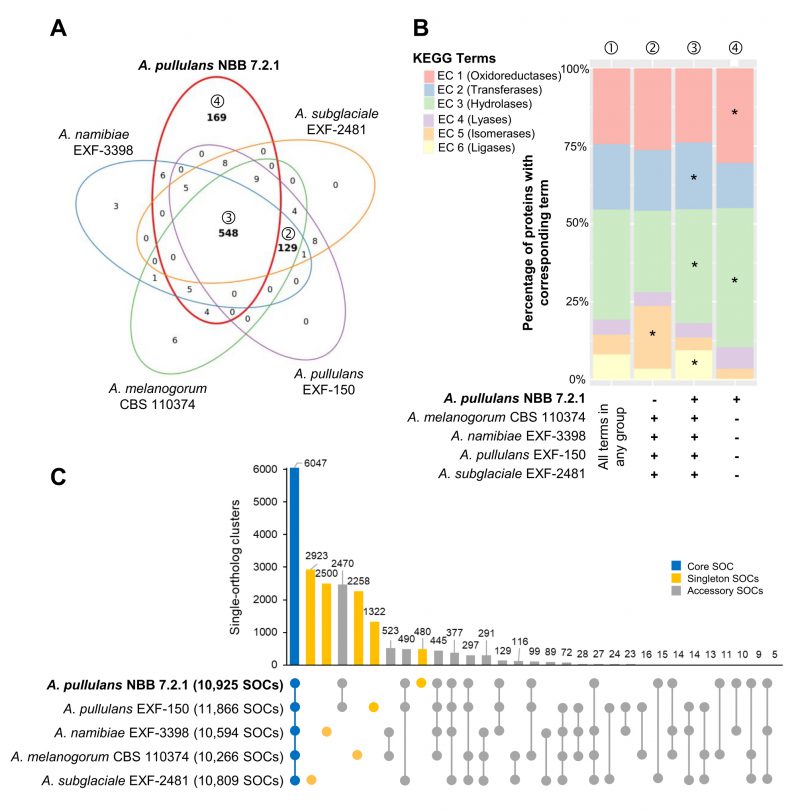
FIGURE 2: Whole genome comparison of five Aureobasidium species. Gene annotations reveal that the A. pullulans NBB 7.2.1 genome contains more unique KEGG terms than the genomes of four other Aureobasidium strains. (A) KEGG term distribution among annotated genes of five Aureobasidium species. 548 terms were commonly found in all species, but a substantial number of terms (169) was unique for the A. pullulans NBB 7.2.1 genome or specifically lacking in this genome (129 terms; all bold). (B) The relative number of genes annotated to one of the six main enzyme classes (EC 1-6; oxidoreductases, transferases, hydrolases, lyases, isomerases, ligases) for all KEGG terms in the five Aureobasidium species and the subsets described in A. The enzyme classes highlighted with an asterisk (*) are significantly overrepresented (adj. p-value≤0.05) in the respective group (as compared to their frequency among all KEGG terms found in any of the five Aureobasidium genomes (①)). Results are shown for those genes with terms shared among all genomes except A. pullulans NBB 7.2.1 (②), shared among all five genomes (③), or only present in A. pullulans NBB 7.2.1 (④). (C) Pan-genome clustering of five Aureobasidium genomes visualized using an Up Set bar diagram [96]. The A. pullulans NBB 7.2.1 genome contained fewer (480) unique gene models than the four other Aureobasidium genomes.
96. Lex A, Gehlenborg N, Strobelt H, Vuillemot R, and Pfister H (2014). UpSet: Visualization of Intersecting Sets. IEEE Trans Vis Comput Graph 20(12): 1983–1992. 10.1109/tvcg.2014.2346248