Back to article: Genome, transcriptome and secretome analyses of the antagonistic, yeast-like fungus Aureobasidium pullulans to identify potential biocontrol genes
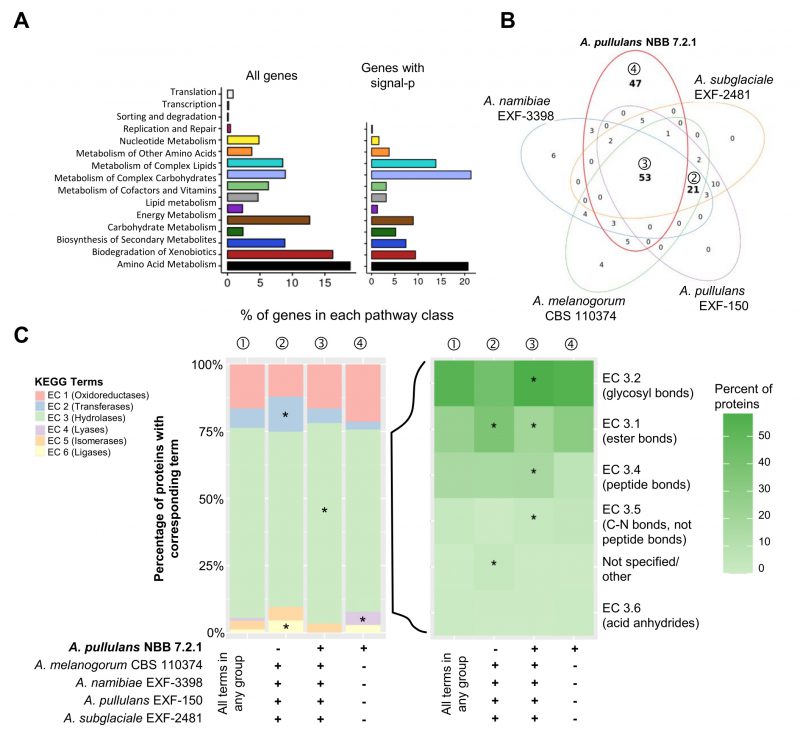
FIGURE 3: Genome mining of Aureobasidium genomes to identify potential biocontrol genes. Functional annotations of the five Aureobasidium genomes (obtained from the DOE-JGI MycoCosm) identified a plethora of genes and gene clusters that may contribute to biocontrol activity. (A) Relative percentage of A. pullulans NBB 7.2.1 genes (for the 4019 genes with a KEGG annotation or only the subset containing a predicted signal peptide) assigned to the different KEGG pathways. (B) KEGG term distribution among the five Aureobasidium genomes for all annotated genes containing a predicted signal peptide. 53 terms were commonly found in all species (③), while 47 terms were unique for the A. pullulans NBB 7.2.1 genome (④) and 21 were exclusively found in the other genomes (②) (bold numbers in Venn diagram). (C) The relative percentage of genes with a predicted signal peptide annotated to belong to one of the six main enzyme classes (EC 1-6; oxidoreductases, transferases, hydrolases, lyases, isomerases, ligases). Results are shown for genes with terms that were found in any of the five genomes (①) and for those shared in all genomes except A. pullulans NBB 7.2.1 (②), shared among all five genomes (③), or only present in A. pullulans NBB 7.2.1 (④). The enzyme classes highlighted with an asterisk were significantly overrepresented (adj. p-value≤0.05) in the respective group. Among the predicted hydrolase genes with a signal peptide, esterases (EC 3.1), glycosylases (EC 3.2), and peptidases (EC 3.4) were by far the most frequent and significantly overrepresented in genes with terms shared by all five Aureobasidium genomes.