Back to article: Genome, transcriptome and secretome analyses of the antagonistic, yeast-like fungus Aureobasidium pullulans to identify potential biocontrol genes
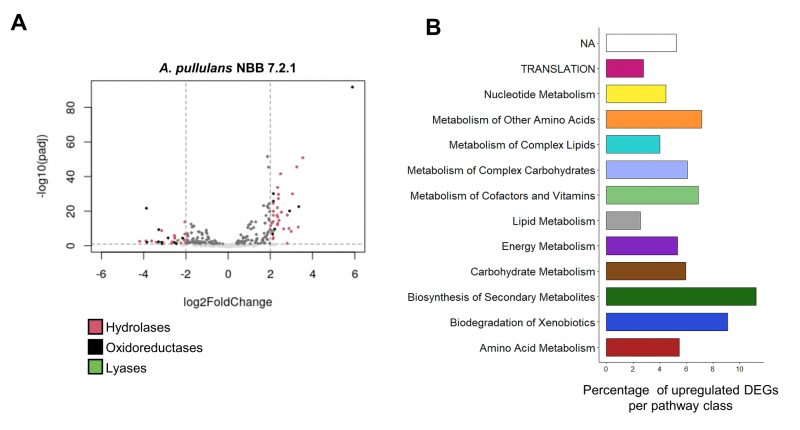
FIGURE 4: Transcriptome analysis of A. pullulans NBB 7.2.1 and F. oxysporum NRRL 26381/CL57 in pure culture and during competition with each other.A. pullulans NBB 7.2.1 strongly responds to co-cultivation with F. oxysporum NRRL 26381/CL57 at the transcriptome level. (A) Volcano plot showing expression of the A. pullulans NBB 7.2.1 genes with a signal peptide. In the co-culture, 178 genes exhibited significantly changed expression by at least a factor of four (92 and 86 up- and downregulated genes, respectively) compared to pure culture. (B) Proportion of upregulated DEGs (log2FoldChange > 2) for genes annotated to different KEGG categories. Biosynthesis of secondary metabolites (11.2%), biodegradation of xenobiotics (9.1%), metabolism of other amino acids (7.1 %), and metabolism of cofactors and vitamins (6.9 %) comprised the categories with the highest frequency of upregulated genes.