Back to article: Genome, transcriptome and secretome analyses of the antagonistic, yeast-like fungus Aureobasidium pullulans to identify potential biocontrol genes
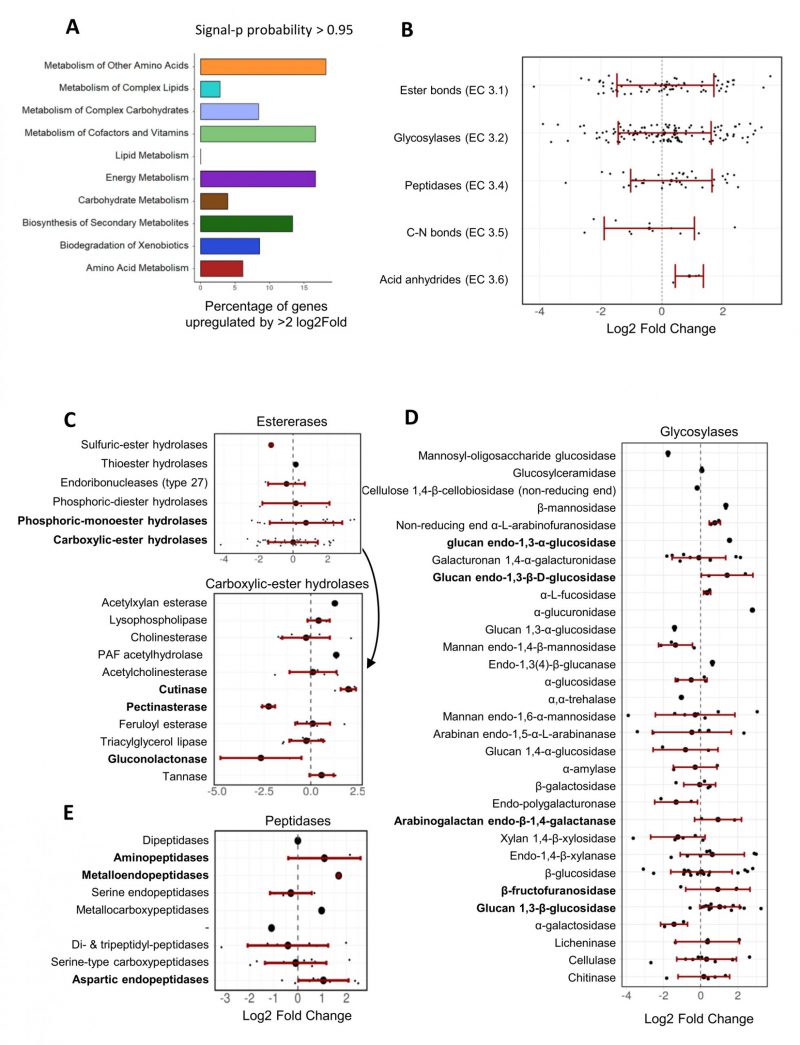
FIGURE 6: Transcriptional regulation of A. pullulans NBB 7.2.1 genes with a predicted signal peptide upon co-cultivation with F. oxysporum NRRL 26381/CL57. Many A. pullulans NBB 7.2.1 hydrolase genes were strongly up- or downregulated during the interaction with F. oxysporum NRRL 26381/CL57. (A) Percentage of upregulated DEGs in each KEGG pathway class for those A. pullulans NBB 7.2.1 genes containing a predicted signal peptide. (B) Variation of the Log2 FC values within each hydrolase enzyme category. Variation of the Log2 FC values of all A. pullulans NBB 7.2.1 DEGs is also shown on a lower level of categorisation for hydrolases acting on ester bonds (C), glycosylases (D), and peptidases (E).