Back to article: Cleavage-defective Topoisomerase I mutants sharply increase G-quadruplex-associated genomic instability
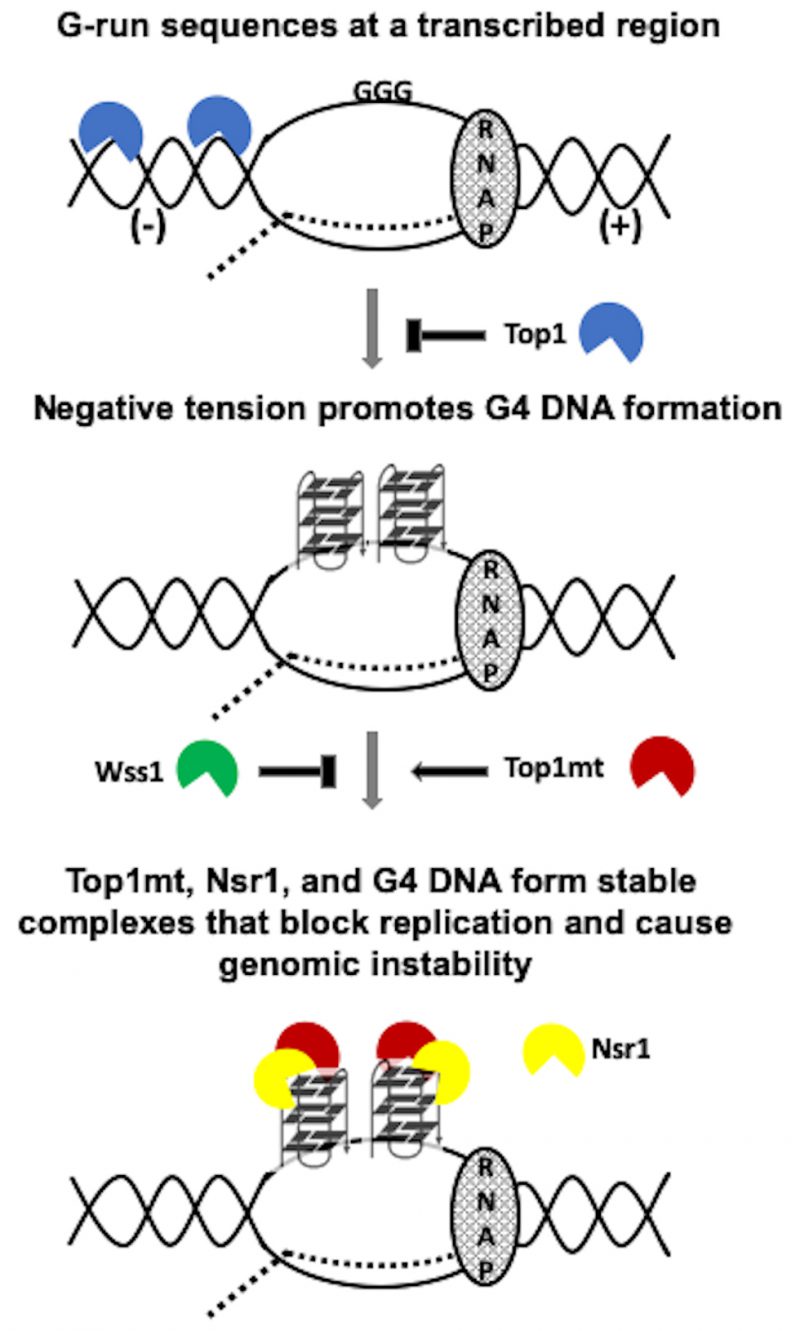
FIGURE 6: Model of co-transcriptional G4-formation and the effect of Top1 activity and mutation on G4-induced genomic instability. RNAP = RNA polymerase complex. Dotted line = the nascent transcript. (-) = negative tension behind the transcription complex. (+) = positive tension ahead of the transcription complex. Top1mt = Top1 mutant. N = Nsr1 N-term. C = Nsr1 C-term. Top1mt capable of G4-binding (i.e. Top1Y727F or Top1Y740Stop) but not Top1mt incapable of G4-binding (i.e. Top1S733E) form Top1mt/Nsr1/G4 DNA complexes that block replication and cause genomic instability.