Research Articles:
Microbial Cell, Vol. 10, No. 11, pp. 248 - 260; doi: 10.15698/mic2023.11.808
Basal level of ppGpp coordinates Escherichia coli cell heterogeneity and ampicillin resistance and persistence
1 Department of Biology, University of Copenhagen, DK-2200 Copenhagen, Denmark.
Keywords: basal ppGpp, resistance, persistence, ampicillin, heterogeneity.
Abbreviations:
ppGpp – guanosine penta and tetra phosphate,
MIC – minimal inhibitory concentration,
PBP – penicillin-binding protein,
RNAP – RNA polymerase,
wt – wild type.
Received originally: 11/05/2023 Received in revised form: 20/09/2023
Accepted: 18/10/2023
Published: 25/10/2023
Correspondence:
Yong Everett Zhang, Department of Biology, University of Copenhagen, DK-2200 Copenhagen, Denmark; yong.zhang@bio.ku.dk
Conflict of interest statement: No conflict of interest is identified with this article.
Please cite this article as: Paulina Katarzyna Grucela and Yong Everett Zhang (2023). Basal level of ppGpp coordinates Escherichia coli cell heterogeneity and ampicillin resistance and persistence. Microbial Cell 10(11): 248-260. doi: 10.15698/mic2023.11.808
Abstract
The universal stringent response alarmone ppGpp (guanosine penta and tetra phosphates) plays a crucial role in various aspects of fundamental cell physiology (e.g., cell growth rate, cell size) and thus bacterial tolerance to and survival of external stresses, including antibiotics. Besides transient antibiotic tolerance (persistence), ppGpp was recently found to contribute to E. coli resistance to ampicillin. How ppGpp regulates both the persistence and resistance to antibiotics remains incompletely understood. In this study, we first clarified that the absence of ppGpp in E. coli (ppGpp0 strain) resulted in a decreased minimal inhibition concentration (MIC) value of ampicillin but, surprisingly, a higher persistence level to ampicillin during exponential growth in MOPS rich medium. High basal ppGpp levels, thus lower growth rate, did not produce high ampicillin persistence. Importantly, we found that the high ampicillin persistence of the ppGpp0 strain is not due to dormant overnight carry-over cells. Instead, the absence of ppGpp produced higher cell heterogeneity, propagating during the regrowth and the killing phases, leading to higher ampicillin persistence. Consistently, we isolated a suppressor mutation of the ppGpp0 strain that restored the standard MIC value of ampicillin and reduced its cell heterogeneity and the ampicillin persistence level concomitantly. Altogether, we discussed the fundamental role of basal level of ppGpp in regulating cell homogeneity and ampicillin persistence.
INTRODUCTION
Bacteria constantly adjust their physiological and metabolic activities and even morphologies in response to unpredictable external environmental cues. Rapid growth rate under non-stressed conditions helps bacteria to compete for nutrients. In contrast, slow growth rate and non-growing dormancy state are essential for bacteria to survive harsh conditions, including starvation, predators, and attacks from the host cell immune systems. Therefore, it is imperative to understand how bacteria dynamically adjust their metabolism to better control them under unwanted conditions, e.g., during bacterial infection.
–
The stringent response alarmone ppGpp (guanosine penta and tetra phosphates) is a universal second messenger that plays a central role in bacterial physiological adaptations [1][2][3][4][5]. Initially discovered under E. coli amino acid starvation conditions [6], ppGpp was later found to be produced under various stress conditions, including the starvation of other nutrients (carbon [7], iron [8], fatty acid [9]). Consistently, ppGpp is essential for stress survival, bacterial virulence, and antibiotic tolerance, persistence, and resistance [1][5][10][11]. The best-studied mechanism is how ppGpp helps E. coli to cope with amino acid starvation by RelA and “ribosome stalling” [12][13][14][15]. During amino acid starvation, the levels of uncharged tRNAs increase, which leads to the stalling of ribosomes on mRNA templates lacking cognate aminoacyl-tRNAs. This ribosome stalling leads to the activation of RelA, which senses the accumulation of uncharged tRNAs and responds by synthesizing ppGpp. ppGpp then binds to the RNA polymerase (RNAP) [16][17] to reprogram global gene expressions, including decreased transcription of genes related to rapid growth and increased expression of genes related to stress response (e.g., amino acid biosynthesis genes), survival, and virulence [18][19][20]. Besides RNAP, ppGpp directly binds and affects the functions of numerous other proteins involved in E. coli translation and metabolism [21][22][23][24]. Altogether, ppGpp, via its numerous target proteins, promptly adjusts bacterial metabolism to adapt to various stresses.
–
Besides in adapting to stressful conditions, the most intriguing role of ppGpp is its connection with antibiotic persistence. Persistence is the phenomenon that a sub-population of isogenic bacterial cells survive the prolonged killing by lethal antibiotics [25][26]. The persistent cells, i.e., persisters, contain no genetic mutation; after antibiotics are removed, persisters regenerate to produce a cell population that undergoes a similar antibiotic killing curve. These phenotypic persisters are thought to originate from noise and heterogeneity of genetic expressions [25][27], can be induced by various stresses [11], and may contribute to recurrent and recalcitrant clinical bacterial infections. Moreover, many studies showed that high persistence facilitates bacterial survival of antibiotic killing and thus more likely leads to the gain of resistance mutations [28][29], thereby multiple drug resistant superbugs.
–
In all studied bacteria, ppGpp is shown to be essential for persistence [11][27][30]. However, other factors, such as the toxin-antitoxin system [31][32][33][34], ATP [35][36], and cell heterogeneity during the lag phase [37][38], are also reported to affect persistence under various conditions. Intriguingly, a recent model proposed by the Groisman group suggests that slow and non-growing states dictate higher levels of persistence in Salmonella [39][40], which seems promising to unify the diverse observations of persistence. Additionally, the same study found that under minimal medium starvation conditions the presence of ppGpp decreases, instead of increases, persistence [39]. The interpretation of these results lies on the fact that ppGpp supports active metabolism and cell growth, rendering them sensitive to antibiotics; while the absence of ppGpp did not allow cell growth, rendering them quiescent to antibiotic actions, leading to higher persistence levels. Notably, ppGpp is a well-established determinant of exponential cell growth rate in E. coli [41]. Artificially increasing ppGpp levels via mutations in SpoT, the other homolog protein of RelA in E. coli, has revealed an inverse relation between ppGpp levels and cell growth rate [41]. Therefore, one would expect a correlation between the level of ppGpp and persistence in exponentially growing non-stressed E. coli cells, which remains to be tested. Therefore, we first aimed to extend the persistence model proposed by Groisman et al. [40] to E. coli, by studying the persistence of strains producing varied basal levels of ppGpp and growing at different rates under non-stressed conditions.
–
Additionally, a new connection between ppGpp and antibiotic resistance was reported [42][43]. A moderate expression of one L,D-transpeptidase YcbB, and simultaneous over-production of ppGpp made E. coli resistant to most β-lactam antibiotics, including ampicillin [43]. This was likely due to the replacement of the 4→3 peptidic cross-links catalyzed by PBPs (penicillin-binding proteins) by the 3→3 cross-links catalyzed by L,D transpeptidases. Since β-lactam antibiotics efficiently inhibit PBPs but not L,D-transpeptidase, using an alternative L,D transpeptidase provided an effective way to resist β-lactams. However, the mechanism of over produced ppGpp in this ampicillin resistance phenomenon remains still unclear. In this regard, previous transcriptomic studies showed that ppGpp influences expression of proteins involved in cell wall metabolism [18][19]. A recent study also showed an inverse correlation between ppGpp levels and cell size of E. coli [44]. In accordance with this, the complete absence of ppGpp in the so-called ppGpp0 strain produces highly filamented E. coli cells [7]. These observations indicate a sophisticated relation between ppGpp, cell division, and ampicillin resistance, persistence.
–
In this study, we found that a basal level of ppGpp is essential for a high MIC value of ampicillin in E. coli. Unexpectedly, we found that exponentially growing cells of the ppGpp0 strain produced a higher level of ampicillin persistent cells than the wild type (wt) strain in MOPS rich medium. Further studies of ppGpp0 and its evolved suppressor strains, and strains with elevated basal ppGpp levels, allowed us to propose that the high cell heterogeneity of the ppGpp0 strain, represented by cell length difference, was responsible for its higher level of persistence towards ampicillin.
RESULTS
The absence of ppGpp reduces the ampicillin MIC value
We first tested the link between ppGpp and ampicillin by measuring the MIC values of the E. coli strains used, i.e., the wt MG1655 strain, the isogenic strain with the relA allele deleted (ΔrelA), or with both the relA deleted and spoT replaced with a chloramphenicol resistance marker (ΔrelA ΔspoT207 strain [7], or the ppGpp0 strain). We chose to use the chemically defined rich MOPS medium supplemented with the twenty amino acids (MOPSr) [45] and 0.2 mM (instead of normally 1.32 mM) phosphate (for short, MOPSr-Lp). This choice was for several reasons. Firstly, the persistence in the LB complex medium varied dramatically due to batch variations of LB; in contrast, MOPSr-Lp contains explicit compositions of nutrients, enhancing data reproducibility. Second, we aimed to investigate the antibiotic responses of E. coli strains growing under non-stressed conditions. MOPSr-Lp provides all the necessary nutrients and trace elements for robust E. coli growth, facilitating our study. Lastly, the low phosphate concentration in MOPSr-Lp offered a robust method for labeling and measuring ppGpp levels with the radioactive isotope 32p-orthophosphate (see below).
–
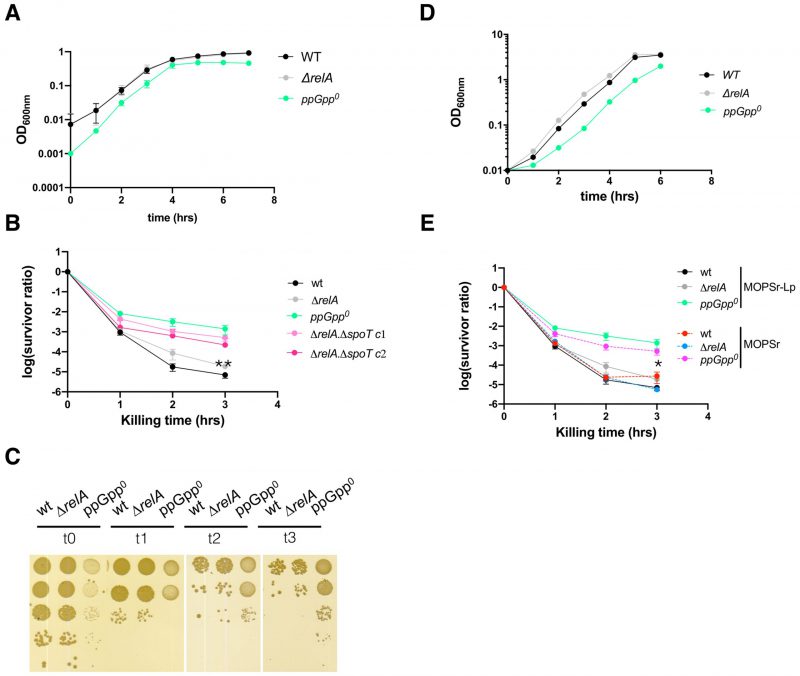
We found that up to 12.5 μg/ml ampicillin did not prevent or slow down the growth or the final yields of both wt and ΔrelA strains in MOPSr-Lp (Figure 1). However, 12.5 μg/ml ampicillin completely inhibited the growth of the ppGpp0 strain for 24 hours, and two-fold serial diluted ampicillin from 12.5 to 1.5625 μg/ml slowed the growth of the ppGpp0 strain in a dose-dependent manner.
–
 |
FIGURE 1: The absence of ppGpp reduces the MIC value of ampicillin in E. coli. (A-D) Growth curves of the three studied strains in MOPS rich medium supplemented with low (0.2 mM) phosphate (MOPSr-Lp) in the presence of two-fold serially diluted ampicillin (from 12.5 to 1.5625 μg/ml). Three biological replicates were performed and the averages and ranges were plotted. WT, E. coli MG1655; ΔrelA, E. coli MG1655 with the relA allele deleted; ppGpp0, ΔrelA ΔspoT207, with spoT replaced with a chloramphenicol resistance marker [7]. |
–
Since the ppGpp0 strain finally grew up after prolonged incubation (72 hrs) in the presence of inhibitory ampicillin, we asked if the ppGpp0 strain obtained any suppressing mutation(s). By re-growing the “evolved” cells in the presence of 6.25 or 3.125 μg/ml ampicillin, we found that they grew up readily within 24 hours and earlier than the original ppGpp0 strain (Figure S1A). This indicates that the ppGpp0 strain obtained some suppressing mutation(s), which allowed it to re-grow faster than the original strain and gain a higher MIC.
–
As controls, we tested two other antibiotics with different modes of action, i.e., gentamycin (targeting translation), and ofloxacin (targeting DNA replication). Despite the slowed growth of the ppGpp0 strain by both 0.125 μg/ml gentamycin and 0.03125 μg/ml ofloxacin (Figure S1B-S1G), the ppGpp0 strain did not obtain any resistance mutation after 72 hours of growth (Figure S1H), suggesting that the ppGpp0 strain has the same MIC values for both gentamycin and ofloxacin as the wt and ΔrelA strains. Altogether, these data showed that the absence of ppGpp reduces the E. coli MIC value and makes it less resistant specifically to ampicillin.
–
The exponential ppGpp0 cells are more persistent to ampicillin killing in MOPSr-Lp
By definition, a lower ampicillin MIC of the ppGpp0 strain indicates that the cell population was not killed but their growth was inhibited by ampicillin. The ppGpp0 cells survived the prolonged ampicillin treatment to eventually evolve out with resistance mutation(s) (Figure S1A), suggesting that the ppGpp0 cells tolerated the sub-MIC ampicillin well. We thus tested if the ppGpp0 strain tolerates a lethal concentration, i.e., 250 μg/ml, ca. 10-20 x MIC of ampicillin, in a persistence assay. To avoid other confounding factors (e.g., stress), we tested exponentially growing cells. For this, 16-hr overnight cultures of the wt, ΔrelA, and the ppGpp0 strains were inoculated into fresh pre-warmed MOPSr-Lp medium and allowed to grow for 2.5 hrs to the early exponential phase (Figure 2A). Ampicillin (250 μg/ml) was then added to kill the cells and the killing kinetics were determined hourly for 3-5 hrs. Surprisingly, the ppGpp0 strain had significantly more persister cells than ΔrelA and wt strains (Figure 2B) and the differences are highly reproducible (six biological replicates, t-test, p=0.011 and p=0.0052 for 3 hr killing time point, respectively). Of note, the colonies of the surviving ppGpp0 cells were smaller, indicative of slower growth and recovery on LB medium (Figure 2C). These small colonies did not contain resistance mutations since a second killing of these colonies in fresh MOPSr-Lp medium produced the same curves as the original ppGpp0 strain (data not shown).
–
–
To further confirm this unexpected phenomenon of the ppGpp0 strain, we re-constructed an in-frame clean deletion mutant of spoT, i.e., ΔrelA ΔspoT, using the scarless method (see Material and Method) [46]. We tested two separate colonies of ΔrelA ΔspoT and found similar killing curves of ampicillin as for the ppGpp0 strain (Figure 2B). In conclusion, both ppGpp0 strains were more persistent to ampicillin than the ΔrelA and wt E. coli strains. We further performed persistence assays for two other antibiotics, i.e., gentamycin and ofloxacin (Figure S2). For gentamycin, we did not observe a significant difference in persistence among the strains. For ofloxacin, we observed a slightly higher level of persistence (<10-fold) in the ppGpp0 strain compared to the ΔrelA strain. This finding suggests that the high persistence and low MIC value observed in the ppGpp0 strain are specific to ampicillin.
–
High ampicillin persistence of ppGpp0 strain holds in standard MOPSr medium
The lower MIC value and higher persistence of the ppGpp0 strains to ampicillin were puzzling. The low phosphate used in the MOPSr-Lp medium may have caused this phenomenon. To test this, the growth dynamics (Figure 2D) and killing assay (Figure 2E) were confirmed in MOPSr supplemented with the standard 1.32 mM phosphate. The persistence level of the wt and ΔrelA strains did not change significantly as compared to those in MOPSr-Lp medium. Despite a small drop of persistence level, the ppGpp0 strain had still significantly more persistent cells than wt and ΔrelA (six biological replicates, t-test, p=0.0258 and p=0.0207 for 3 hr killing time point, respectively). This argues that the high persistence of the ppGpp0 strain cannot be attributed to the low phosphate in MOPSr-Lp.
–
A lower growth rate did not dictate higher ampicillin persistence in MOPSr-Lp
The ppGpp0 strain has a slightly lower growth rate (doubling time 0.54 ± 0.02 hr) than the wt and relA strains (0.48 ± 0.02 vs. 0.48 ± 0.03 hr, respectively, Figure 3A), which might cause the higher persistence to ampicillin according to the proposed model [39][40]. Additionally, the ppGpp level dictates the E. coli growth rate [41]. To test if a higher basal ppGpp level, thus a lower growth rate, leads to higher persistence, we employed strains of E. coli K-12 carrying point mutations in spoT known to produce higher basal levels of ppGpp. These strains included the SpoT202 (T78I) [47] and SpoT203 (R140C) [47]. Isogenic MG1655 strains carrying the mutations were reported to produce 7- and 4-times increased levels of ppGpp as compared to wt. SpoT is essential when RelA is present in MG1655, and relA is dispensable for cell growth in rich MOPSr medium with twenty amino acids. To avoid potential influence from the residual RelA activity and simplify the genetic engineering of spoT on the chromosome, we introduced these SpoT mutations in the endogenous spoT allele in the ΔrelA deletion strain of MG1655 by using the scar-less method.
–
 |
FIGURE 3: High basal ppGpp and thus low growth rate did not produce more ampicillin persistent cells. (A) Growth dynamics of the spoT202 and spoT203 strains. For comparison, the growth curves of the wt, ΔrelA and ppGpp0 strains from Figure 2A were included. Four biological replicates were performed, and the averages and SDs are shown. spoT202, ΔrelA spoT (T78I) [47]; and spoT203, ΔrelA spoT (R140C) [47]. (B) Thin layer chromatography analysis of the intracellular ppGpp levels in the five exponentially growing strains as labeled with 32p-orthophosphate in the MOPSr-Lp medium. GTP, ppGpp and pppGpp are indicated. Unknown molecules nearby the ppGpp are framed in broken-line rectangles. (C) Quantification of the ppGpp levels as in panel (B). Two biological replicates were performed, and the average and SD are shown. (D) The ampicillin (250 μg/ml) killing curves of the studied strains. Three biological replicates of both spoT202 and spoT203 were performed, and the averages and SDs are shown. For comparison, the same killing curves of the three strains from (Figure 2B) were included. |
–
We found that wt MG1655 re-grew from a 16-hr overnight culture as early as the strains ΔrelA, followed by the ppGpp0 strain, then spoT202 and spoT203 (Figure 3A). These lead to varied doubling times of the strains, with spoT202 and spoT203 exhibiting a growth rate that was half of the wt, ΔrelA and the ppGpp0 strains (doubling time 1.02 ± 0.05 and 0.99 ± 0.08 hr, respectively). These data confirm that 1) RelA is not required for the growth of E. coli in MOPSr-Lp; 2) that the expected high level of ppGpp in spoT202 and spoT203 reduced their growth rates significantly [47]. To corroborate, we measured the intracellular ppGpp with the radioactive 32p-orthophosphate (Figure 3B) [21]. As expected, the exponentially growing cells of both spoT202 and spoT203 strains produced higher ppGpp levels than the wt strain, which is higher than both ΔrelA and the ppGpp0 strains with very low, if any, basal levels of ppGpp (Figure 3C).
–
With the spoT mutant strains constructed and their ppGpp levels confirmed, we then sought to use them to study the effect of higher basal ppGpp levels and, thus slower growth rate on ampicillin persistence. Surprisingly we found that both the spoT202 and spoT203 strains were rapidly killed by ampicillin, and after three hours, both had similar persistence levels to that of the wt strain (Figure 3D). These data demonstrated that higher basal levels of ppGpp and thus lower growth rates do not lead to higher ampicillin persistence in exponentially growing cells in MOPSr-Lp.
–
Overnight carry-over cells did not fully account for the high persistence of ppGpp0 strain
Both wt MG1655 and ΔrelA re-grew rapidly and earlier than the ppGpp0 strain in MOPSr-Lp (Figure 3A). A more extended lag phase is known to contain a subpopulation of cells in a dormant state that avoids the killing by antibiotics. If true, one would expect to see proportionally more persistent cells when more overnight cells are inoculated. To test this, we increased the inoculum by five or ten times and tested the persistence again (Figure S3). We found that the persistent cells of the ppGpp0 strain increased by ca. 100 x and 1000 x fold, respectively, when the inoculum increased by 5 x and 10 x fold (Figure 4A, 4B). On the other hand, the persistent cells of both wt and ΔrelA strains increased less dramatically (10-100 x fold). As controls, the overnight cells were completely resistant to the killing, consistent with the fact that ampicillin kills actively growing and dividing cells (Figure 4C). Therefore, despite that the increased persistent cells of all three strains can be partially from the overnight dormant cells, another factor contributes significantly more to ampicillin persistence during regrowth, and such a persistence “amplification” effect is more pronounced in the ppGpp0 strain than in wt and ΔrelA strains.
–
–
We hypothesized that the high ampicillin persistence of the ppGpp0 strain could be caused by a higher cell heterogeneity during regrowth. Subculturing cells one more time should reduce both dormant cells, lag time, and cell heterogeneity and, thus the persistence level. To test this, we subcultured the strains first for 3 hrs from OD600nm 0.01, then subcultured the cells again from OD600nm 0.01 for 2.5 hrs before subjecting them to killing. We observed a drop in persistence of all three strains, but the ppGpp0 strain still produced ten times more persistent cells than the wt and ΔrelA strains (Figure 4D, 3 hr time point). This result indicates that the exponentially growing ppGpp0 cells produce high cell heterogeneity in MOPSr-Lp, leading to high persistence to ampicillin.
–
High cell heterogeneity of the ppGpp0 strain during regrowth and ampicillin killing
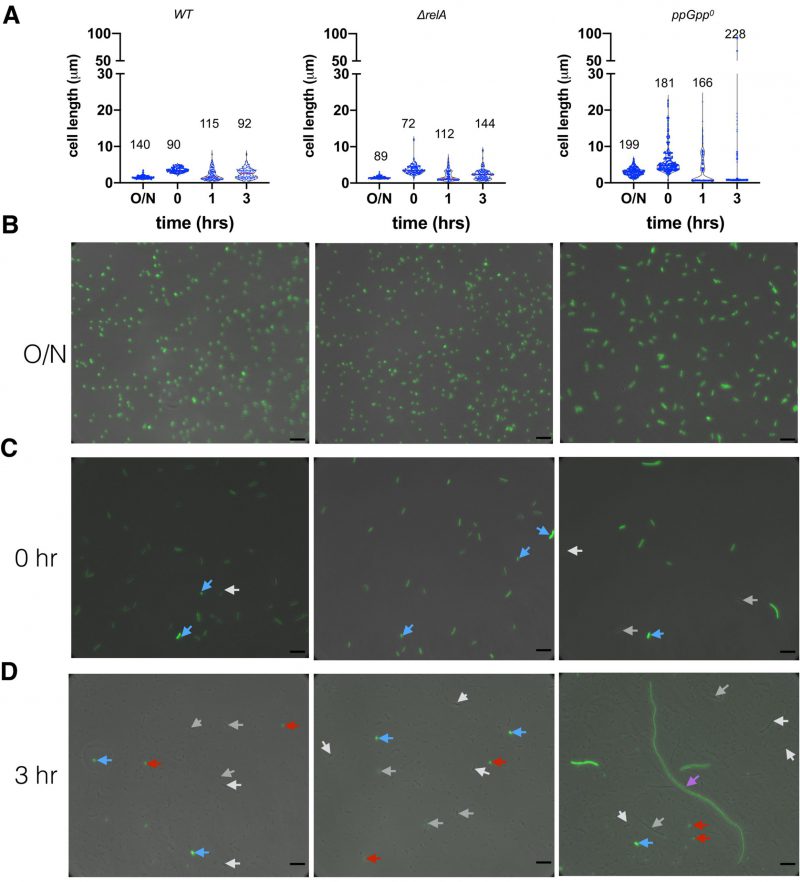
The above data suggest that the heterogeneity of the ppGpp0 strain leads to more persistent cells to ampicillin during regrowth in the rich MOPS medium. To corroborate this, we constructed the p7 GFP gene [48] under the pBAD promoter of pBAD33 and transformed the plasmid into the three strains. Arabinose was added into the pre-cultures in MOPSr-Lp to express the p7 GFP. The arabinose was then washed away to stop further expression of p7 GFP. These cells were used to inoculate fresh MOPSr-Lp, and similar growth dynamics and ampicillin killing curves were observed, indicative of no major effect from p7GFP expression (data not shown). The exponential phase cells of these strains were subjected to ampicillin killing, and cells were inspected at different time points under a fluorescence microscope (Figure 5).
–
–
For stationary phase cells, we observed E. coli cells with characteristic short rods for both wt and ΔrelA strains (cell length 1.57 ± 0.46 µm, N=140, and 1.55 ± 0.37 µm, N=89, respectively; student t-test p=0.733). The ppGpp0 cells, however, were significantly longer (3.09 ± 1.09 µm, N=199, student t-test p <0.0001 between ppGpp0 and ΔrelA), consistent with previous report [7]. Further, the ppGpp0 cells showed a higher heterogeneity in terms of cell length (Figure 5A, 5B; SD/average ratio 0.35 of ppGpp0 vs. 0.24 of ΔrelA) [49], suggesting a role of basal ppGpp in regulating cell length and size homogeneity. All cells of the three strains showed a strong green fluorescence of p7 GFP (Figure 5B). Upon regrowth to the exponential phase (time point 0, Figure 5C), both wt and ΔrelA cells elongate to 3.56 ± 0.69 µm and 3.74 ± 1.4 µm with good uniformity (SD/average ratio 0.19 and 0.37), while a more significant cell length heterogeneity of ppGpp0 was observed (6.15 ± 3.9 µm, SD/average ratio 0.63) as in [7]. Some ppGpp0 cells are 12-22 µm long, which was not observed for the wt and ΔrelA strains. Note that the varied dim green fluorescence intensity in cells indicates a dilution of the stable p7 GFP through cell divisions (white and grey arrows, Figure 5C). We also noticed potential overnight carry-over dormant cells with strong fluorescence intensity (blue arrows, Figure 5C).
–
The 3-hr killing by ampicillin led to an even higher cell-length heterogeneity of all strains, because of the production of round, highly fluorescent, thus likely dying, mini cells (average diameter ca. 0.65 µm; Figure 5D, red arrows). The wt and ΔrelA cells did not elongate during ampicillin killing, and fluorescence was dim for most of the surviving cells (Figure 5A, 5D, white and grey arrows), suggesting they are offspring cells after several rounds of cell division. The ppGpp0 strain also produced dim surviving cells with intermediate length (ca. 15 µm; white and grey arrows). Remarkably, some humongous, elongated cells (magenta arrow, Figure 5D) of ppGpp0 were observed, which were ca. 90 µm long, thus 4-15 times longer than normal time zero ppGpp0 cells (Figure 5C). These cells emitted weak green fluorescence as compared to the bright dormant cells (blue arrow), indicating that they were overnight cells growing out of the stationary phase. However, their significantly increased cell length indicates that these cells were growing in MOPSr-Lp despite the presence of a lethal concentration of ampicillin. Although dormant bright cells (blue arrows) were found for all three strains, we believe that the dim and elongated growing cells of ppGpp0 likely contributed to its significantly more persistent cells to ampicillin.
–
The ppGpp0 suppressing mutants with rescued ampicillin MIC become less heterogeneous and persistent
The ppGpp0 strain has lower ampicillin MIC (Figure 1), and the suppressing mutants (hereinafter called ppGpp0-AS) frequently occurred (Figure S1A). We isolated several ppGpp0-AS strains after 72 hrs of growth of the strain ΔrelA ΔspoT (YZ841) in the presence of ampicillin (12.5 μg/ml). We found that the tested five ppGpp0-AS strains all grew up in the presence of ampicillin and thus restored the ampicillin MIC to the standard value of wt and ΔrelA strains (Figure 6A), indicating stable mutation(s) were present in these strains. We also found that these ppGpp0-AS strains (YZ1094, YZ1097) became less persistent to ampicillin and behaved like the wt and ΔrelA strains (Figure 6B). Moreover, the suppressing strains showed rescued heterogeneity in cell length during both regrowth and ampicillin killing as compared to the parental strain ΔrelA ΔspoT (YZ841) (Figure 6C).
–
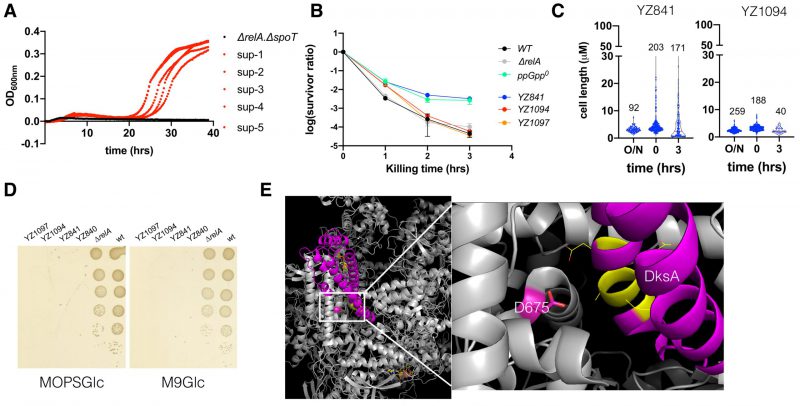 |
FIGURE 6: The ppGpp0 suppressing mutants rescued the ampicillin MIC and are less heterogeneous and persistent. (A) Growth curves of the parental ΔrelA ΔspoT strain and the five suppressor strains in the presence of 12.5 μg/ml ampicillin. (B) The ampicillin (250 μg/ml) killing curves of the parental ΔrelA ΔspoT strain (YZ841) and the two suppressor strains (YZ1094, YZ1097). Three biological replicates were performed, and the average and SEM were presented. For comparison, the same killing curves of the three strains from (Figure 2B) were included. (C) Box plot of the cell length (µm) of the parental ΔrelA ΔspoT strain and the suppressor strain YZ1094 cells during regrowth and ampicillin killing. The numbers of analyzed cells are indicated above the plots. (D) Growth of the parental ΔrelA ΔspoT(YZ840/YZ841) and the suppressor strains (YZ1094/YZ1097) on MOPS (left) and M9 (right) minimal media agar plate supplemented with glucose 0.2% g/ml (MOPSGlc and M9Glc). Pictures were taken after 24 hrs growth at 37°C. (E) 3D position of the suppressing mutation D675A on RpoB, in a complex structure of E. coli RNAP (grey), DksA (magenta), and two molecules of ppGpp (yellow stick model) (PDB 5VSW) [50]). D675 is shown in a stick model (magenta). To the right side is a zoom-in of the framed region, with DksA residues directly facing D675 shown in yellow and line model. |
–
It is known that the ppGpp0 strain frequently obtains mutations during the stringent response. To check if the ppGpp0-AS strains contain a stringent mutation, we tested their growth on MOPS and M9 minimal media supplemented with glucose 0.2% (g/ml) but without amino acids. Figure 6D shows that, like the parental strains, none of the ppGpp0-AS strains grew on either medium, while the positive control strains wt and ΔrelA did. These data suggest that the suppressing mutations encoded in these ppGpp0-AS strains are not stringent. To identify the mutation(s), we first amplified the rpoB gene of the two ppGpp0-AS strains since rpoB is a mutation hot spot. Sanger sequencing revealed that an A2024C mutation occurred on both YZ1094 and YZ1097 strains, leading to a D675A amino acid change. D675 is a highly conserved residue and confronts the backbone of DksA residue Alanine77 (PDB 5VSW, [50]; Figure 6E). Further whole genome sequencing revealed no other mutation besides A2024C, suggesting that D675A was responsible for restoring the typical MIC of ampicillin, homogenous cell length, and reduced persistence to ampicillin of the ppGpp0 strain.
DISCUSSION
In this study, we found that the absence of ppGpp reduced the MIC value of ampicillin in E. coli and, surprisingly, that exponentially growing cells of the ppGpp0 strain are more persistent to ampicillin than the wt and ΔrelA strains. Further experiments showed that a higher basal level of ppGpp and thus lower growth rate did not lead to high ampicillin persistence under non-stressed conditions (Figure 3). Instead, we found that the high persistence levels to ampicillin cannot be (solely) attributed to slow or absence of cell growth, or overnight carry-over cells, but rather was due to a high cell heterogeneity, represented by cell length/size (Figures 4, 5). Many studies showed that high antibiotic persistence facilitates bacterial gain of resistance mutations [28][29]. Significantly, we readily isolated a suppressor mutation of ppGpp0 strains, which concomitantly restored the lower MIC value, and reduced its cell homogeneity and the persistence level to ampicillin (Figure 6), strongly arguing for a critical role of basal levels of ppGpp in regulating cell heterogeneity and ampicillin persistence and resistance.
–
The lower ampicillin MIC of the ppGpp0 strain is consistent with other reports; however, the high ampicillin persistence of exponentially growing ppGpp0 strain appears to contradict with many other studies. We suspect that several reasons may account for this discrepancy. 1) We used MOPSr chemically defined rich medium, while many other studies used LB complex medium. 2) We made and used the E. coli K-12 MG1655 strain and its isogenic strains with in-frame clean deletions or mutations of relA and spoT in the endogenous chromosomal loci. This minimizes unnecessary heterogeneity derived from e.g., gene dosage difference from plasmid vectors, residual antibiotic marker of the e.g., KEIO collection strains [51]. 3) We studied the exponential phase cells, which are not expected to experience significant stress except for the varying levels of basal ppGpp (Figure 3). This allowed us to directly test the hypothesis that basal ppGpp levels dictate the persistence level. Indeed, the persister cells we studied can be viewed as the spontaneous (type II) persisters generated by non-stressed cells, which have been rarely studied [25], whereas, most other persistence studies involve cells experiencing some stress conditions that generate type I persisters [11]. These stresses include, for instance, nutrient starvation, low pH, low oxygen, and host cell immune system. Under these stressful conditions, ppGpp firstly is essential for transcribing genes responsible for bacterial coping with these stresses [5]. The absence of ppGpp thus predisposed bacteria to these stresses, further confounding the study of its role on persistence. 4) Lastly, the high ampicillin persistence phenomenon was observed before in exponential phase cells of E. coli ppGpp0 strain [11]. Furthermore, we tested two different ppGpp0 strains and six biological replicates produced highly consistent results (Figure 2), suggesting that this phenomenon was authentic.
–
How to interpret the high ampicillin persistence of the ppGpp0 strain growing under non-stressed conditions? The ppGpp0 strain of E. coli was known many years ago to form long filamentous cells [7]. Consistently, we found increasing heterogeneity of exponential cells from the wt, ΔrelA to the ppGpp0 strains (Figure 5A). This agrees with the gradually increased cell size (and heterogeneity) of E. coli when the level of ppGpp was tuned down from near stringent to sub-basal levels [44]. In accordance, we observed rapid killing by ampicillin of the wt strain, intermediate killing of ΔrelA and the slowest killing of the ppGpp0 strain (Figure 2B). Furthermore, the strains spoT202 and spoT203 contain higher levels of basal ppGpp but they are less persistent to ampicillin (Figure 3). We propose that both mutants with higher basal levels of ppGpp exhibit greater control over cell size heterogeneity that is comparable to wt cells, but is higher than the ΔrelA strain, and finally the ppGpp0 strain. Conversely, wt cells exhibited a persistence level similar to that of both mutants, while the ΔrelA strain displayed higher persistence, with the highest level being observed in the ppGpp0 strain. In total, it appears that under non-stressed exponentially growing conditions, basal levels of ppGpp are inversely corelated with cell heterogeneity and ampicillin persistence.
–
Despite the correlation, the underlying mechanism of how ppGpp regulates cell size heterogeneity remained elusive. In this study, we obtained random suppressors of the ppGpp0 cells following exposure to sub-MIC levels of ampicillin (Figure 6). These suppressors not only restored a normal ampicillin MIC value but also led to a simultaneous reduction in both cell heterogeneity and ampicillin persistence. These observations strongly suggest a pivotal role of basal ppGpp in orchestrating cell growth, division, and, consequently, cell size heterogeneity and ampicillin persistence. To explore this correlation further, we attempted to analyze the gene expression differences among ampicillin-persistent cells from wt, ΔrelA, and ppGpp0 strains using bulk RNA sequencing. However, due to the significant variability in the samples, likely stemming from the high heterogeneity of persister cells, we were unable to draw definitive conclusions from these data (not shown). Instead, we believe single persister cell sorting and single-cell RNA sequencing may hold the potential. Despite this, many similar analyses were performed in E. coli cells under stringent conditions, thus providing a picture of how stringent, high level of ppGpp controls the transcription of individual genes. For example, Traxler et al. probed transcriptomic differences between wt MG1655 and a ppGpp0 mutant during isoleucine starvation [19]. Fifteen genes involved in cell division significantly differed between the two strains, with nine up-regulated (i.e., yibP/envC, tig, gidA, ftsX, ftsP, cedA, obgE, nlpI, rrmJ) and six down-regulated (i.e., xerC, tolC, yjbQ, bolA, fic, dacC/pbp6) in the ppGpp0 strain. Importantly, Uehara et al. showed that four LytM domain proteins, including EnvC, are essential to activate amidases for segregating daughter cells [52]. Indeed, the absence of the four LytM domain proteins (ΔenvC ΔnlpD ΔygeR ΔyebA) made E. coli cells elongate and less sensitive to ampicillin killing [52]. Besides, endopeptidases such as MepS and MepM are essential for 3→3 based peptidoglycan network expansion during cell growth [42]. Moreover, the outer membrane lipoprotein NlpI potentially interacts with murein endopeptidases PBP4/1, PBP7, MepS, and MepM [53]. Altogether, given the effect of ppGpp on gene expression of multiple cell wall related enzymes, it is conceivable that some dysregulation of cell wall growth and cell division occurs in the ppGpp0 strain, thereby leading to the enhanced cell size heterogeneity.
–
How does then the higher cell size heterogeneity contribute to high ampicillin persistence of ppGpp0 cells? One possible explanation is that ppGpp0 cells are predisposed to hyper-respond to ampicillin given the proposed dysregulated gene expression pattern [19] and thereby increase the persistence level [40]. However, we observed actively growing cells of ppGpp0 during the ampicillin-killing phase (Figure 5D), indicating that it was not cell dormancy which prevented their killing by ampicillin. Instead, the morphological change of ppGpp0 cells during both regrowth and ampicillin killing phase suggest dramatic physiological changes that either pre-exist or are exaggerated further by ampicillin treatment, or more likely, both. Cell lysis rates induced by β-lactams correlate with bacterial growth rate [54] and, importantly, cell lysis requires a functional cell division machinery, the divisome [55]. The filamented cells of the ppGpp0 strain indicate a malfunctional divisome, which may somehow link to its higher persistence, similar as the quadruple mutant of LysM domain proteins reported in [52]. Further, E. coli cells killed by ampicillin are known to elongate during the process, generating a characteristic cell morphology that can be used to distinguish modes of action of different antibiotics [56]. Using these experimental data, recent mathematical modeling took a step further and suggested that a lower surface-to-volume (S/V) ratio of E. coli growing under non-stressed condition are beneficial for antibiotic resistance [57]. In this scenario, elongated cells have a lower S/V ratio, thus a lower influx rate of antibiotics, while rich medium supports fast growth, thus producing ample antibiotic targets and a fast dilution of effective antibiotics. ppGpp0 cells are naturally longer during regrowth in non-stressed condition (Figure 5C); meanwhile, rich MOPS medium supports active growth of ppGpp0 cells even during ampicillin killing, as estimated from the highly elongated cells (Figure 5D). Altogether, it is conceivable that the ppGpp0 cells have malfunctional divisomes that lead to high cell heterogeneity, which potentially serve as a bet-hedge strategy to avoid ampicillin killing. Despite these plausible connections, a mechanistic study is highly warranted to explicitly uncover the underlying mechanism of ppGpp in regulating cell heterogeneity and high ampicillin persistence and resistance.
MATERIALS AND METHODS
Bacterial strains, growth, media, and antibiotics
The constructed strains and primers used in this project are listed in Table S1 and S2, respectively. The E. coli K-12 MG1655 is the wild type strain used. Lysogeny broth (LB, containing 10 g tryptone (Oxoid), 5 g yeast extract (Oxoid), and 10 g NaCl (SIGMA) per liter of distilled water) was the primary medium used for E. coli growth and cloning. The defined MOPS rich medium (MOPSr) is the same as in [45]. The defined MOPS rich medium with low phosphate is the same as MOPSr, except that 0.2 mM, instead of 1.32 mM, K2HPO4 is supplemented. The MOPSGlc medium was the same as MOPSr, except that no amino acid was supplemented. M9 minimal medium (M9Glc) contains Na2HPO4 48 mM, KH2PO4 22 mM, pH 7.4, NaCl 8.6 mM, NH4Cl 18.7 mM, MgSO4 1 mM, CaCl2 0.1 mM, thiamine 1 μg/ml, glucose 0.2% g/ml. When applicable, 1.5% g/ml agar is added to make agar plates. Chloramphenicol 25 μg/ml, ampicillin 100 μg/ml, and anhydrotetracycline were used for the scarless cloning process.
–
Scarless chromosomal mutagenesis of spoT
To introduce a point mutation in the spoT gene on the chromosome, the scar-less mutagenesis method reported in [46] was used. Briefly, primers PYZ395/396 were used to amplify the SceI-Cam fragment from the pWRG100 plasmid, which was electroporated into ΔrelA containing pWRG99 (YZ313), generating strain YZ580. The primer pairs PYZ397/P71 and PYZ398/P69 were then used to amplify the two DNA fragments containing the T78I mutation, which were later fused via overlap PCR by using primers P69/P71. This DNA was electroporated into YZ580 to replace the SceI-Cam fragment and obtain the spoT202(T78I) strain. Similarly, for strain spoT203(R140C), primer pairs PYZ400/P71 and PYZ399/P69 were used to amplify the two DNA fragments containing the R140C mutation, which were fused together by using primers P69/P71. These DNAs were used to obtain the spoT203(R140C) strain. To make a clean in-frame deletion of spoT, oligo PYZ351 was used to replace the SceI-Cam fragment of strain YZ580. The respective spoT mutant sequences of the constructed strains were confirmed by PCR amplification and Sanger sequencing before use.
–
Measurements of ppGpp by autoradiography
ppGpp were measured and quantitated the same as described in [24]. Two biological replicates were performed, and the average and SD were presented.
–
MIC measurement
Overnight pre-cultures of the strains in MOPSr-Lp medium were washed with fresh MOPSr-Lp medium and inoculated (initial OD600nm=0.025, ca. CFU 2 x 10^7) into 200 μl fresh MOPSr-Lp medium supplemented with serially diluted antibiotics (starting concentrations of gentamycin 0.25 μg/ml, ofloxacin 0.125 μg/ml, ampicillin 25 μg/ml). The strains were allowed to grow in microtiter plates (LABSOLUTE, no. 7696794) for 72 hrs at 30°C with vigorous agitation (548 cpm, double orbit) in the Biotek Synergy H1 plate reader. Three biological replicates were performed.
–
Selection of ppGpp0 suppressors in the presence of ampicillin
Strain YZ841 was grown in MOPSr-Lp medium supplemented with 12.5 μg/ml ampicillin for 72 hrs at 30°C with vigorous agitation (548 cpm, double orbit) in the Biotek Synergy H1 plate reader. Cells were washed with PBS to remove residual ampicillin before plating on an LB agar plate and incubated for 24 hrs at 37°C. Colonies (ca. 20) were picked and tested with their growth in MOPSr-Lp medium supplemented with 12.5 μg/ml ampicillin for 24 hrs, with the parental strain YZ841 as a negative control. Five candidate suppressors were selected that showed active growth within 24 hrs, as compared to the parental strain YZ841.
–
Ampicillin killing assay
The killing assay was performed similarly as described [24]. Briefly, 16-hr overnight cultures of each strain were made in two ml MOPSr-Lp medium in a snap-tube (Sarstedt, no. 62.515.006). Approximately 10^6-2 x 10^6 cells of each strain were inoculated into 125 ml flasks containing 10 ml fresh MOPSr-Lp medium pre-warmed at 37°C. Cells were grown in a water bath at 37°C with agitation (160 rpm). After 2.5 hours of growth, lethal concentrations of ampicillin (250 µg/ml) were added in the cultures to kill the cells for 3 hrs or up to 5/8 hrs at 37°C with agitation (160 rpm). Right before antibiotic exposure, the total CFUs were determined by serial dilution, spotting, and incubation at 37°C for 24 hrs before counting. After each hour of antibiotic killing, 1.4 ml of cells were removed, spun down at 5000 rpm 3 min and then 14000 rpm 2 min. Cells were washed once with 1 ml of PBS, spun down as above, and finally resuspended in PBS. The CFUs of persister cells were determined by serial dilution, spotting and incubating on LB agar plate, and counting as above.
–
Fluorescence microscopy
Bacterial cells out of overnight growth, regrowth in fresh MOPSr-Lp medium, and after different hours of ampicillin killing were visualized under a Nikon Eclipse Ti-E inverted microscope equipped with a 100x, 1.45 NA objective, a Zyla 5.5 sCMOS camera and a NIS-ELEMENTS software. All cells were washed with and resuspended in the M9Glc medium before imaging. Fiji and plugin MicrobeJ software [58] were used for automatic bacterial cell detection and quantification.
–
Whole genome resequencing
Whole genome resequencing of the suppressor strains was performed at Genewiz and the sequence was analyzed and mutations identified using Artemis [59].
–
Statistical analysis
GraphPad Prism v.8 was used throughout to analyze the data.
REFERENCES
- Hauryliuk V, Atkinson GC, Murakami KS, Tenson T, Gerdes K (2015). Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat Rev Microbiol 13(5): 298-309. 10.1038/nrmicro3448
- Fernandez-Coll L, Cashel M (2020). Possible Roles for Basal Levels of (p)ppGpp: Growth Efficiency Vs. Surviving Stress. Front Microbiol 11: 592718. 10.3389/fmicb.2020.592718
- Irving SE, Choudhury NR, Corrigan RM (2021). The stringent response and physiological roles of (pp)pGpp in bacteria. Nat Rev Microbiol 19(4): 256-271. 10.1038/s41579-020-00470-y
- Chau NYE, Ahmad S, Whitney JC, Coombes BK (2021). Emerging and divergent roles of pyrophosphorylated nucleotides in bacterial physiology and pathogenesis. PLoS Pathog 17(5): e1009532. 10.1371/journal.ppat.1009532
- Dalebroux ZD, Svensson SL, Gaynor EC, Swanson MS (2010). ppGpp conjures bacterial virulence. Microbiol Mol Biol Rev 74(2): 171-199. 10.1128/MMBR.00046-09
- Cashel M, Gallant J (1969). Two compounds implicated in the function of the RC gene of Escherichia coli. Nature 221(5183): 838-841. 10.1038/221838a0
- Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M (1991). Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. Journal Biol Chem 266(9): 5980-5990. 10.1016/s0021-9258(19)67694-5
- Vinella D, Albrecht C, Cashel M, D’Ari R (2005). Iron limitation induces SpoT-dependent accumulation of ppGpp in Escherichia coli. Mol Microbiol 56(4): 958-970. 10.1111/j.1365-2958.2005.04601.x
- Seyfzadeh M, Keener J, Nomura M (1993). spoT-dependent accumulation of guanosine tetraphosphate in response to fatty acid starvation in Escherichia coli. Proc Natl Acad Sci USA 90(23): 11004-11008. 10.1073/pnas.90.23.11004
- Pacios O, Blasco L, Bleriot I, Fernandez-Garcia L, Ambroa A, Lopez M, Bou G, Canton R, Garcia-Contreras R, Wood TK, Tomas M (2020). (p)ppGpp and Its Role in Bacterial Persistence: New Challenges. Antimicrob Agents Chemother 64(10): e01283-20. 10.1128/AAC.01283-20
- Harms A, Maisonneuve E, Gerdes K (2016). Mechanisms of bacterial persistence during stress and antibiotic exposure. Science 354(6318): aaf4268. 10.1126/science.aaf4268
- Winther KS, Roghanian M, Gerdes K (2018). Activation of the Stringent Response by Loading of RelA-tRNA Complexes at the Ribosomal A-Site. Mol Cell 70(1): 95-105 e104. 10.1016/j.molcel.2018.02.033
- Arenz S, Abdelshahid M, Sohmen D, Payoe R, Starosta AL, Berninghausen O, Hauryliuk V, Beckmann R, Wilson DN (2016). The stringent factor RelA adopts an open conformation on the ribosome to stimulate ppGpp synthesis. Nucleic Acids Res 44(13): 6471-6481. 10.1093/nar/gkw470
- Brown A, Fernandez IS, Gordiyenko Y, Ramakrishnan V (2016). Ribosome-dependent activation of stringent control. Nature 534(7606): 277-280. 10.1038/nature17675
- Loveland AB, Bah E, Madireddy R, Zhang Y, Brilot AF, Grigorieff N, Korostelev AA (2016). Ribosome*RelA structures reveal the mechanism of stringent response activation. Elife 5: e17029. 10.7554/eLife.17029
- Ross W, Sanchez-Vazquez P, Chen AY, Lee JH, Burgos HL, Gourse RL (2016). ppGpp Binding to a Site at the RNAP-DksA Interface Accounts for Its Dramatic Effects on Transcription Initiation during the Stringent Response. Mol Cell 62(6): 811-823. 10.1016/j.molcel.2016.04.029
- Ross W, Vrentas CE, Sanchez-Vazquez P, Gaal T, Gourse RL (2013). The magic spot: a ppGpp binding site on E. coli RNA polymerase responsible for regulation of transcription initiation. Mol Cell 50(3): 420-429. 10.1016/j.molcel.2013.03.021
- Sanchez-Vazquez P, Dewey CN, Kitten N, Ross W, Gourse RL (2019). Genome-wide effects on Escherichia coli transcription from ppGpp binding to its two sites on RNA polymerase. Proc Natl Acad Sci USA 116(17): 8310-8319. 10.1073/pnas.1819682116
- Traxler MF, Summers SM, Nguyen HT, Zacharia VM, Hightower GA, Smith JT, Conway T (2008). The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol Microbiol 68(5): 1128-1148. 10.1111/j.1365-2958.2008.06229.x
- Durfee T, Hansen AM, Zhi H, Blattner FR, Jin DJ (2008). Transcription profiling of the stringent response in Escherichia coli. J Bacteriol 190(3): 1084-1096. 10.1128/JB.01092-07
- Zhang Y, Zbornikova E, Rejman D, Gerdes K (2018). Novel (p)ppGpp Binding and Metabolizing Proteins of Escherichia coli. Mbio 9(2): e02188-17. 10.1128/mBio.02188-17
- Wang B, Dai P, Ding D, Del Rosario A, Grant RA, Pentelute BL, Laub MT (2019). Affinity-based capture and identification of protein effectors of the growth regulator ppGpp. Nat Chem Biol 15(2): 141-150. 10.1038/s41589-018-0183-4
- Wang B, Grant RA, Laub MT (2020). ppGpp Coordinates Nucleotide and Amino-Acid Synthesis in E. coli During Starvation. Mol Cell 80(1): 29-42 e10. 10.1016/j.molcel.2020.08.005
- Zhang YE, Baerentsen RL, Fuhrer T, Sauer U, Gerdes K, Brodersen DE (2019). (p)ppGpp Regulates a Bacterial Nucleosidase by an Allosteric Two-Domain Switch. Mol Cell 74(6): 1239-1249 e1234. 10.1016/j.molcel.2019.03.035
- Balaban NQ, Helaine S, Lewis K, Ackermann M, Aldridge B, Andersson DI, Brynildsen MP, Bumann D, Camilli A, Collins JJ, Dehio C, Fortune S, Ghigo JM, Hardt WD, Harms A, Heinemann M, Hung DT, Jenal U, Levin BR, Michiels J, Storz G, Tan MW, Tenson T, Van Melderen L, Zinkernagel A (2019). Definitions and guidelines for research on antibiotic persistence. Nat Rev Microbiol 17(7): 441-448. 10.1038/s41579-019-0196-3
- Brauner A, Fridman O, Gefen O, Balaban NQ (2016). Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat Rev Microbiol 14(5): 320-330. 10.1038/nrmicro.2016.34
- Urbaniec J, Xu Y, Hu Y, Hingley-Wilson S, McFadden J (2022). Phenotypic heterogeneity in persisters: a novel ‘hunker’ theory of persistence. FEMS Microbiol Rev 46(1). 10.1093/femsre/fuab042
- Levin-Reisman I, Ronin I, Gefen O, Braniss I, Shoresh N, Balaban NQ (2017). Antibiotic tolerance facilitates the evolution of resistance. Science 355(6327): 826-830. 10.1126/science.aaj2191
- Levin-Reisman I, Brauner A, Ronin I, Balaban NQ (2019). Epistasis between antibiotic tolerance, persistence, and resistance mutations. Proc Natl Acad Sci USA 116(29): 14734-14739. 10.1073/pnas.1906169116
- Verstraete L, Van den Bergh B, Verstraeten N, Michiels J (2022). Ecology and evolution of antibiotic persistence. Trends Microbiol 30(5): 466-479. 10.1016/j.tim.2021.10.001
- Moyed HS, Bertrand KP (1983). hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J Bacteriol 155(2): 768-775. 10.1128/jb.155.2.768-775.1983
- Wang X, Wood TK (2011). Toxin-antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl Environ Microbiol 77(16): 5577-5583. 10.1128/AEM.05068-11
- Verstraeten N, Knapen WJ, Kint CI, Liebens V, Van den Bergh B, Dewachter L, Michiels JE, Fu Q, David CC, Fierro AC, Marchal K, Beirlant J, Versees W, Hofkens J, Jansen M, Fauvart M, Michiels J (2015). Obg and Membrane Depolarization Are Part of a Microbial Bet-Hedging Strategy that Leads to Antibiotic Tolerance. Mol Cell 59(1): 9-21. 10.1016/j.molcel.2015.05.011
- Dorr T, Vulic M, Lewis K (2010). Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. Plos Biol 8(2): e1000317. 10.1371/journal.pbio.1000317
- Shan Y, Brown Gandt A, Rowe SE, Deisinger JP, Conlon BP, Lewis K (2017). ATP-Dependent Persister Formation in Escherichia coli. Mbio 8(1): e02267-16. 10.1128/mBio.02267-16
- Kwan BW, Valenta JA, Benedik MJ, Wood TK (2013). Arrested protein synthesis increases persister-like cell formation. Antimicrob Agents Chemother 57(3): 1468-1473. 10.1128/AAC.02135-12
- Fridman O, Goldberg A, Ronin I, Shoresh N, Balaban NQ (2014). Optimization of lag time underlies antibiotic tolerance in evolved bacterial populations. Nature 513(7518): 418-421. 10.1038/nature13469
- Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S (2004). Bacterial persistence as a phenotypic switch. Science 305(5690): 1622-1625. 10.1126/science.1099390
- Pontes MH, Groisman EA (2019). Slow growth determines nonheritable antibiotic resistance in Salmonella enterica. Sci Signal 12(592): eaax3938. 10.1126/scisignal.aax3938
- Pontes MH, Groisman EA (2020). A Physiological Basis for Nonheritable Antibiotic Resistance. Mbio 11(3): e00817-20. 10.1128/mBio.00817-20
- Potrykus K, Murphy H, Philippe N, Cashel M (2011). ppGpp is the major source of growth rate control in E. coli. Environ Microbiol 13(3): 563-575. 10.1111/j.1462-2920.2010.02357.x
- Voedts H, Dorchene D, Lodge A, Vollmer W, Arthur M, Hugonnet JE (2021). Role of endopeptidases in peptidoglycan synthesis mediated by alternative cross-linking enzymes in Escherichia coli. EMBO J 40(19): e108126. 10.15252/embj.2021108126
- Hugonnet JE, Mengin-Lecreulx D, Monton A, den Blaauwen T, Carbonnelle E, Veckerle C, Brun YV, van Nieuwenhze M, Bouchier C, Tu K, Rice LB, Arthur M (2016). Factors essential for L,D-transpeptidase-mediated peptidoglycan cross-linking and beta-lactam resistance in Escherichia coli. Elife 5: e19469. 10.7554/eLife.19469
- Buke F, Grilli J, Cosentino Lagomarsino M, Bokinsky G, Tans SJ (2022). ppGpp is a bacterial cell size regulator. Curr Biol 32(4): 870-877 e875. 10.1016/j.cub.2021.12.033
- Neidhardt FC, Bloch PL, Smith DF (1974). Culture medium for enterobacteria. J Bacteriol 119(3): 736-747. 10.1128/jb.119.3.736-747.1974
- Blank K, Hensel M, Gerlach RG (2011). Rapid and highly efficient method for scarless mutagenesis within the Salmonella enterica chromosome. PLoS One 6(1): e15763. 10.1371/journal.pone.0015763
- Sarubbi E, Rudd KE, Cashel M (1988). Basal ppGpp level adjustment shown by new spoT mutants affect steady state growth rates and rrnA ribosomal promoter regulation in Escherichia coli. Mol Gen Genet 213(2-3): 214-222. 10.1007/BF00339584
- Fisher AC, DeLisa MP (2008). Laboratory evolution of fast-folding green fluorescent protein using secretory pathway quality control. PLoS One 3(6): e2351. 10.1371/journal.pone.0002351
- Lapinska U, Voliotis M, Lee KK, Campey A, Stone MRL, Tuck B, Phetsang W, Zhang B, Tsaneva-Atanasova K, Blaskovich MAT, Pagliara S (2022). Fast bacterial growth reduces antibiotic accumulation and efficacy. Elife 11: e74062. 10.7554/eLife.74062
- Molodtsov V, Sineva E, Zhang L, Huang X, Cashel M, Ades SE, Murakami KS (2018). Allosteric Effector ppGpp Potentiates the Inhibition of Transcript Initiation by DksA. Mol Cell 69(5): 828-839 e825. 10.1016/j.molcel.2018.01.035
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H (2006). Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2: 2006 0008. 10.1038/msb4100050
- Uehara T, Dinh T, Bernhardt TG (2009). LytM-domain factors are required for daughter cell separation and rapid ampicillin-induced lysis in Escherichia coli. J Bacteriol 191(16): 5094-5107. 10.1128/JB.00505-09
- Banzhaf M, Yau HC, Verheul J, Lodge A, Kritikos G, Mateus A, Cordier B, Hov AK, Stein F, Wartel M, Pazos M, Solovyova AS, Breukink E, van Teeffelen S, Savitski MM, den Blaauwen T, Typas A, Vollmer W (2020). Outer membrane lipoprotein NlpI scaffolds peptidoglycan hydrolases within multi-enzyme complexes in Escherichia coli. EMBO J 39(5): e102246. 10.15252/embj.2019102246
- Tuomanen E, Cozens R, Tosch W, Zak O, Tomasz A (1986). The rate of killing of Escherichia coli by beta-lactam antibiotics is strictly proportional to the rate of bacterial growth. J Gen Microbiol 132(5): 1297-1304. 10.1099/00221287-132-5-1297
- Chung HS, Yao Z, Goehring NW, Kishony R, Beckwith J, Kahne D (2009). Rapid beta-lactam-induced lysis requires successful assembly of the cell division machinery. Proc Natl Acad Sci USA 106(51): 21872-21877. 10.1073/pnas.0911674106
- Nonejuie P, Burkart M, Pogliano K, Pogliano J (2013). Bacterial cytological profiling rapidly identifies the cellular pathways targeted by antibacterial molecules. Proc Natl Acad Sci USA 110(40): 16169-16174. 10.1073/pnas.1311066110
- Ojkic N, Serbanescu D, Banerjee S (2022). Antibiotic Resistance via Bacterial Cell Shape-Shifting. Mbio 13(3): e0065922. 10.1128/mbio.00659-22
- Ducret A, Quardokus EM, Brun YV (2016). MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat Microbiol 1(7): 16077. 10.1038/nmicrobiol.2016.77
- Carver T, Harris SR, Berriman M, Parkhill J, McQuillan JA (2012). Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 28(4): 464-469. 10.1093/bioinformatics/btr703
–
SUPPLEMENTAL INFORMATION
![]() Download Supplemental Information
Download Supplemental Information
ACKNOWLEDGMENTS
This study was supported by an Novo Nordisk Foundation Project Grant (NNF19OC0058331) to Y.E.Z.
COPYRIGHT
© 2023

Basal level of ppGpp coordinates Escherichia coli cell heterogeneity and ampicillin resistance and persistence by Grucela and Zhang is licensed under a Creative Commons Attribution 4.0 International License.