Research Articles:
Microbial Cell, Vol. 4, No. 2, pp. 38 - 51; doi: 10.15698/mic2017.02.556
Balanced CoQ6 biosynthesis is required for lifespan and mitophagy in yeast
Centro Andaluz de Biología del Desarrollo, Universidad Pablo de Olavide-CSIC, CIBERER Instituto de Salud Carlos III, Sevilla, 41013, Spain.
Keywords: coenzyme Q6, regulation, mitochondria, yeast, mitophagy, chronological life span.
Received originally: 19/07/2017 Received in revised form: 29/11/2016
Accepted: 19/01/2017
Published: 03/02/2017
Correspondence:
C. Santos-Ocaña, csanoca@upo.es
Conflict of interest statement: The authors declare no conflict of interest.
Please cite this article as: Isabel González-Mariscal, Aléjandro Martín-Montalvo, Cristina Ojeda-González, Adolfo Rodríguez-Eguren, Purificación Gutiérrez-Ríos, Plácido Navas, and Carlos Santos-Ocaña (2017). Balanced CoQ6 biosynthesis is required for lifespan and mitophagy in yeast. Microbial Cell 4(2): 38-51.
Abstract
Coenzyme Q is an essential lipid with redox capacity that is present in all organisms. In yeast its biosynthesis depends on a multiprotein complex in which Coq7 protein has both catalytic and regulatory functions. Coq7 modulates CoQ6 levels through a phosphorylation cycle, where dephosphorylation of three amino acids (Ser/Thr) by the mitochondrial phosphatase Ptc7 increases the levels of CoQ6. Here we analyzed the role of Ptc7 and the phosphorylation state of Coq7 in yeast mitochondrial function. The conversion of the three Ser/Thr to alanine led to a permanently active form of Coq7 that caused a 2.5-fold increase of CoQ6 levels, albeit decreased mitochondrial respiratory chain activity and oxidative stress resistance capacity. This resulted in an increase in endogenous ROS production and shortened the chronological life span (CLS) compared to wild type. The null PTC7 mutant (ptc7∆) strain showed a lower biosynthesis rate of CoQ6 and a significant shortening of the CLS. The reduced CLS observed in ptc7Δ was restored by the overexpression of PTC7 but not by the addition of exogenous CoQ6. Overexpression of PTC7 increased mitophagy in a wild type strain. This finding suggests an additional Ptc7 function beyond the regulation of CoQ biosynthesis. Genetic disruption of PTC7 prevented mitophagy activation in conditions of nitrogen deprivation. In brief, we show that, in yeast, Ptc7 modulates the adaptation to respiratory metabolism by dephosphorylating Coq7 to supply newly synthesized CoQ6, and by activating mitophagy to remove defective mitochondria at stationary phase, guaranteeing a proper CLS in yeast.
INTRODUCTION
Coenzyme Q (CoQ) deficiency is a syndrome that belongs to the family of mitochondrial diseases [1]. CoQ, a lipid embedded in cell membranes, main function is to act as an electron carrier and as an antioxidant. CoQ deficiency is classified by the observed phenotype as it is a multiple-caused syndrome [2]. There are primary and secondary CoQ deficiencies; the deficiency can be a consequence of mutations in genes involved directly in the CoQ biosynthesis (primary deficiency) [3][4] or a consequence of defects or mutations in genes not directly related to CoQ biosynthesis (secondary deficiency) [5][6]. The existence of secondary deficiencies introduces the idea of regulatory mechanisms of CoQ biosynthesis that could also be related to a general regulation of mitochondrial metabolism. The main defect caused by CoQ deficiency is a depletion of ATP in tissues. The variety of symptoms and the diversity of CoQ functions introduce a source of complexity in the analysis of CoQ deficiency syndrome that requires the analysis of the mechanisms regulating the biosynthesis of CoQ [7].
–
In yeast, CoQ6 is synthesized in the mitochondria after the condensation of an activated polyisoprenoid tail with a hydroxybenzoic ring; the ring is further modified by several reactions [8]. The enzymes that catalyze these reactions are encoded by nuclear COQ genes [9]. Most of the COQ genes encode proteins responsible for enzymatic activity; however, three proteins without enzymatic activity, Coq4, Coq8 and Coq9, play a structural role. Coq4 has been reported to support the assembly of the CoQ6 biosynthetic complex in yeast [10]. Coq8 has been included in a family of unusual kinases [11], which also includes other proteins involved in CoQ biosynthesis [12]. Recent analysis of the function of human ADCK3 protein, homologous of yeast Coq8, showed that the inhibition of ADCK3 kinase activity is required for the activation of CoQ6 biosynthesis [13][14]. Coq9 is a membrane protein located at the matrix side of the mitochondrial inner membrane and it belongs to the CoQ6 biosynthetic complex, where it co-migrates with Coq3 and Coq4 at a molecular mass of approximately 1 MDa [15] and it binds to Coq7 to promote CoQ6 biosynthesis [16].
–
Yeast biosynthesis of CoQ6 occurs in a multi-protein complex (Q-synthome). The assembly of the Q-synthome requires the post-translational modification of Coq proteins. Several studies in the last years have demonstrated the existence of the Q-synthome [17][18][19] and several models for the assembly of the complex have been proposed [9][20]. The complex assembly starts with a nucleation around the quinone-like lipid polyprenyl benzoate bound to a nucleating Coq protein such as Coq4. The nucleation step is ended with the assembly of a pre-complex that accumulates a CoQ6 intermediate, the demethoxy quinone (DMQ6) [19][21]. DMQ6 is converted to CoQ6 after the activation of Coq7 by dephosphorylation [22]. Coq7 catalyzes the next to last reaction of the pathway [23], the DMQ6 hydroxylation. Several studies have reported the existence of phosphoproteins in the family of Coq proteins: Coq3, Coq5 and Coq7 [24][25], but only phosphorylation of Coq7 is known to have a physiological relevance [22]. Coq7 phosphorylation leads to a low activity state, therefore accumulating DMQ6, while its dephosphorylation activates Coq7 and increases CoQ6 levels. Both activation states of Coq7 can be achieved by changing the carbon source in the culture media [22]. These results were confirmed in COQ7 null mutants yeast strains (coq7∆) expressing either a Coq7 version that is permanently dephosphorylated (Coq7-AAA), associated with a sharp increase of CoQ6 concentration, or a permanently phosphorylated version (Coq7-DED) that is associated with a significant decrease of CoQ6 levels [22]. The Coq7-AAA version was obtained by site-directed mutagenesis of residues S20, S28 and T32 to alanine, while in the Coq7-DED version these residues were mutated to glutamic or aspartic acid. Recent studies have demonstrated the presence of another phosphorylatable residue in Coq7, the S114 [26], whose modification affects the catalytic function of Coq7. The activation (i.e. dephosphorylation) of Coq7 is carried out by Ptc7, a phosphatase that belongs to the type 2C Mg2+ or Mn2+ dependent protein phosphatases, PPM [27][28]. Other mitochondrial members of this family (Ptc5) have been related to the activation of pyruvate dehydrogenase (PDH) [29][30] and with the activation of mitophagy (Ptc6) [31][32]. Null mutants of PTC7 gene (ptc7∆) have decreased levels of CoQ6, decreased mitochondrial respiratory chain activities and decreased resistance to oxidative stress [28]. The expression of Coq7-AAA in ptc7∆ did not disrupt the high amount of CoQ6 produced by this Coq7 version, demonstrating a relationship between Coq7 and Ptc7 [28].
–
Here we investigate the effect of both Coq7 mutants, Coq7-AAA and Coq7-DED on mitochondrial physiology and its relationship with Ptc7 function. Our results indicate that although Coq7 mutants modify the mitochondrial physiology in a similar fashion than ptc7∆ relative to CoQ6 levels, they have different effects on chronological life span, respiratory complexes interactions and on mitophagy activation.
RESULTS
Coq7-phosphorylation mutants show defects in mitochondrial respiratory chain activities
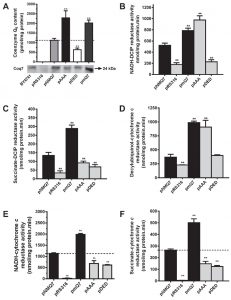
We measured CoQ6 content and the activities of mitochondrial respiratory chain (MRC) in single or coupled complexes (Figure 1). The strain coq7∆ did not contain CoQ6, which was rescued in the control strain (coq7∆/pNMQ7). The strain expressing permanently dephosphorylated Coq7 (coq7∆/pAAA) showed a dramatic increase of CoQ6, while the strain expressing permanently phosphorylated Coq7 (coq7∆/pDED) shows a significant decrease of CoQ6 compared to control. Multicopy COQ7 transformed yeast (coq7∆/pmQ7) also significantly increased CoQ6 (Figure 1A). Under the experimental conditions and in all the strains analyzed, Coq7 was detected by western blotting at the expected size of 24 kDa. NADH-Q reductase activity, measured as NADH-DCIP reductase, was decreased in the coq7∆/pDED strain in a similar manner than negative control (coq7∆/pRS316), but in the coq7∆/pAAA strain, the activity was increased over the control, equivalent to the activity measured in the coq7∆/pmQ7 strain (Figure 1B). Complex II activity measured as succinate-DCIP reductase showed a moderated decrease in both coq7∆/pDED and coq7∆/pAAA strains (Figure 1C). However, complex II activity was increased significantly in the coq7∆/pmQ7 strain. Complex III activity (decylubiquinol-cytochrome c reductase) showed changes comparable to those in complex I (Figure 1D). Coupled MRC activities require CoQ6 as electron carrier, which is not added exogenously in the assay. Complexes activities such as NADH-cytochrome c reductase and succinate-cytochrome c reductase (Figures 1E and 1F) were decreased in both coq7∆/pAAA and coq7∆/pDED strains compared to control. Interestingly, the decrease in NADH-cytochrome c reductase activity in the coq7∆/pAAA strain does not correlate with the increased activity observed in single complexes (Figure 1B and 1D) and with the high amount of CoQ6 found in mitochondria of the coq7∆/pAAA strain. The activities of these complexes in the coq7∆/pmQ7 strain were significantly higher than in control.
Oxidative stress conditions in Coq7 phosphomutants
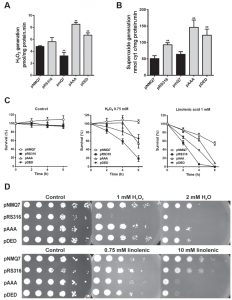
Due to the changes observed in MRC, we analyzed the endogenous oxidative stress, measured as H2O2 generation in mitochondria, from coq7∆ yeast expressing the different versions of Coq7 (Figure 2A). Expression of both Coq7-pAAA and Coq7-pDED showed an increased generation of H2O2 in mitochondria compared to control, while the coq7∆/pmQ7 strain showed a decreased amount, even lower than control. To determine the oxidative stress produced specifically by the complex III, we analyzed the su-peroxide anion generation using reduced decylubiquinone and acetylated cytochrome c (Figure 2B) [33]. Strains expressing both mutated versions of Coq7 produced significantly higher amounts of superoxide, from 200 to 400%, compared to wild type. Also, superoxide was higher in the coq7∆/pRS316 strain. On the contrary, the coq7∆/pmQ7 strain showed a superoxide production comparable to control.
In yeast, CoQ6 acts as a powerful antioxidant and it is required to protect against lipophilic oxidants such as linolenic acid [34][35][36]. The sensitivity to either H2O2 or linolenic acid was analyzed in a coq7∆ strain expressing both Coq7 versions harboring phosphosite modifications (Figures 2C-D). Survival was not compromised in all strains without oxidative stress insult (Figure 2C) but after incubation with H2O2 or linolenic acid the wild type strain showed the higher survival rate at 2, 4 and 6 hours, while the negative control (coq7∆/pRS316) showed a lower survival rate. Interestingly, both treatments in coq7∆/pAAA and coq7∆/pDED strains produced a similar effect on survival, being higher than negative control but lower than control. A similar effect was found when the assay was performed in agar plates with H2O2 (Figure 2D); the lack of CoQ6 in the coq7∆/pRS316 strain compromised the growth but coq7∆/pAAA and coq7∆/pDED growth was only slightly lower compared to control. When the assay was performed with linolenic acid at higher concentration, both coq7 mutants, coq7∆/pAAA and coq7∆/pDED, were more sensitive to the treatment, although both retain CoQ6 production. That result is surprising since null Coq mutants do not produce CoQ6 and show a high sensitivity to linolenic acid [35][36]. In contrast, coq7∆/pAAA and coq7∆/pDED strains synthesize CoQ6, with higher production than control for coq7∆/pAAA strain.
–
Chronological life span is differentially affected in Coq7 phosphomutants
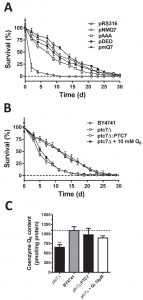
Chronological lifespan (CLS) measurement refers to yeast cells longevity in an exhausted culture media after the onset of stationary phase [37]. The extension of CLS depends on the existence of an intact respiratory metabolism [38][39][40][41] and also by the expression of intact antioxidant defenses [42][43][44]. Given that these requirements are affected in cells expressing both Coq7-pAAA and Coq7-pDED, CLS have been measured in these cells (Figure 3A and Table S1). The coq7∆/pRS316 strain showed shorter mean CLS (2.8 ± 0.2 days) compared to both coq7∆/pNMQ7 (12.2 ± 0.7 days) and coq7∆/pmQ7 strains (14 ± 0.8 days). The coq7∆/pDED strain showed a slightly shorter mean CLS (11.4 ± 0.8 days) while the coq7∆/pAAA strain had a clearly shorter mean CLS (9.1 ± 0.7 days). These results compared to CoQ6 content (Figure 1A) support a model where the phosphorylation state of Coq7 affects CLS in yeast. Comparing the levels of CoQ6 and CLS in the Coq7 versions we found an assortment of results. Thus, CoQ6 level alone is not a factor that can explain the changes observed in CLS.
Ptc7 regulates CoQ6 biosynthesis by dephosphorylation of Coq7 [28]. We studied whether the lack of Ptc7 would result in a compromised CLS. The ptc7∆ strain displayed a shortened mean CLS compared to wild type (6.8 ± 0.4 days versus 12.7 ± 0.7 days) (Figure 3B and Table S2). These results indicated that this phosphatase might regulate the normal function of longevity-associated pathways in yeast. CoQ6 content of the ptc7∆ strain was significantly lower than both wild type and ptc7∆ strains rescued with the wild type allele (Figure 3C). As a result, CoQ6 supplementation in CoQ6 deficient coq7∆ yeast, rescues respiratory growth and oxidative stress resistance [45]. This was demonstrated by measuring the CLS of our strains in CoQ6 supplemented media (Figure 3B). Interestingly, the addition of exogenous CoQ6 to the ptc7∆ strain increased mitochondrial CoQ6 to wild type levels (Figure 3C) but did not rescue CLS of the strain (7 ± 0.8 days), suggesting that the decreased CoQ6 content is not responsible for the shortened CLS of the ptc7∆ strain. These results indicate that CoQ6 levels cannot explain alterations of CLS in both coq7∆ and ptc7∆ mutants. Moreover, the reduction in CLS in the ptc7∆ strain indicates that there might be additional functions of Ptc7 besides Coq7 activation.
–
Respiratory supercomplexes are altered in Coq7 phosphomutants
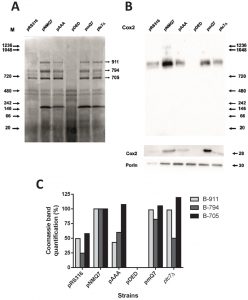
Mutant versions of Coq7 showed a clear defect on MRC activities focused in coupled reactions (NADH-cytochrome c reductase and succinate-cytochrome c reductase) that cannot be explained by the levels of CoQ6. In the coq7∆/pAAA strain the NADH-cytochrome c reductase activity is 40% less than wild type while the simple activity of both separated complexes is even higher than wild type. However, coq7∆/pAAA mitochondria contain 250% of CoQ6 compared to wild type (Figure 1). One possibility is that the expression of Coq7 versions can modify the stability or assembly of respiratory complexes. To this end, we analyzed the assembly of respiratory complexes by BN-PAGE in permeabilized mitochondria isolated from coq7∆ yeast expressing several versions of Coq7 (Figure 4). Cells were cultured first in YPD to increase cell mass and then the culture was transferred to YPG to activate mitochondrial metabolism. The coq7∆/pNMQ7 strain showed a typical profile of mitochondrial supercomplexes that mostly agrees with previously published data in wild type yeast [46], with three prominent bands at 911, 794 and 705 kDa, a double band around 480 kDa and a band located at 242 kDa (Figure 4A). The mass-spectrometry analysis of the three larger bands of coq7∆/pNMQ7 yeast indicated that the 911 kDa band corresponds mainly to complex V, the band of 794 kDa to complex III + complex IV and 705 KDa correspond to complex V. The detailed list of detected and identified proteins can be consulted at the Table S3 of Supplementary Material. Full MASCOT analysis is included in Supplementary Material. Immunoblot of this BN-PAGE gel with anti-Cox2 antibody (Figure 4B) showed differential intensities of supercomplex at 794 kDa but not in other bands. No reaction was observed in mitochondria from coq7∆/pDED yeast, and very low in mitochondria from coq7∆/pRS316 yeast. Lower intensities were also found in both coq7∆/pAAA and ptc7∆ strains. Higher intensity was observed in control coq7∆/pNMQ7 and coq7∆/pmQ7 strains. The quantitative analysis of Coomassie staining showed that the 911 kDa and 794 kDa bands were significantly affected in both Coq7 mutants (Figure 4C). The expression of modified versions of Coq7 induced alterations in the assembly profile of respiratory complexes, being more dramatic in the coq7∆/pDED strain.
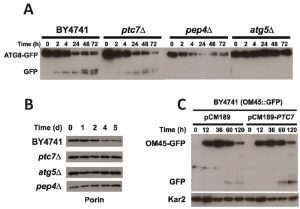
Mitophagy but not autophagy is affected in ptc7∆ mutant strains
Mitophagy is a general process that degrades damaged or non-useful mitochondria, which is required to maintain CLS in eukaryotic cells [31][47][48]. The ptc7∆ strain combines a low amount of CoQ6 with a low CLS while the coq7∆/pDED strain shows a low amount of CoQ6 but a CLS comparable to wild type (Figures 1A and 3A). Both strains are equivalent in terms of Coq7 phosphorylation state and therefore Ptc7 must have another function independent of CoQ6 biosynthesis that is responsible for the low CLS measured. We have previously demonstrated that the survival of human fibroblasts deficient in CoQ10 production depends on a proper recycling of dysfunctional mitochondria by mitophagy [49]. We speculated that Ptc7 might regulate this process. Therefore, we analyzed whether the general process of autophagy and/or mitophagy could be compromised in ptc7∆. Macroautophagy was analyzed by monitoring the Atg8p proteolysis using a plasmid expressing Atg8-GFP tag at the C-terminal under endogenous promoter (ATG8-GFP) (Figure 5A) [50]. Yeast cells were grown in YPD for 16 hours and subsequently were resuspended in nitrogen deprived medium (SDc–N) to induce macroautophagy. The positive control strain (BY4741; wild type) resulted in a marked increase of Atg8-GFP degradation, visible as free-GFP starting at 2 hours, which was maintained until 72 hours. According to previous reports, pep4∆ and atg5∆ strains, which are deficient in protein degradation in the vacuole or autophagy respectively, showed impaired autophagy induction [51] (Figure 5A). In accordance with induced autophagy, a decrease of intact Atg8-GFP and a subsequent increase on free GFP was also observed in ptc7∆ strain, indicating that macroautophagy induction is not compromised in this strain.
To further study the potential role of Ptc7 in mitochondrial recycling, mitophagy was analyzed by monitoring the degradation of porin. Yeasts were grown in YPL for 16 hours and media was replaced with a nitrogen deprived media (SDc–N) to induce mitophagy (Figure 5B). The wild type strain showed a decreased amount of porin starting at day 2 and was gradually reduced until day 5. Porin degradation was not observed in the ptc7∆ strain under similar conditions, suggesting that Ptc7 is involved in mitochondrial recycling. Porin degradation was not observed in the autophagy-deficient yeast strains atg5∆ and pep4∆. A protein expression analysis normalized with total protein loaded corroborated previous data (Figure 2S). To further determine the involvement of Ptc7 in mitophagy induction we determined the effect of Ptc7 overexpression on mitophagy (Figure 5C). Wild type yeast harboring the GFP protein fused in frame with the mitochondrial outer membrane protein Om45 were transformed with the empty yeast expression plasmid (pCM189) or containing the yeast PTC7 coding sequence (pCM189-PTC7). Yeast cells were cultured in YPD for 16 hours and media was replaced with YPL, a non-fermentable carbon source, to induce mitochondrial biosynthesis. Yeast cells were grown through prolonged stationary phase to induce the selective recycling of mitochondria by mitophagy [31]. PTC7 over-expression produced increased GFP free levels starting at 60 hours (270%) and at 120 hours (470%) of growth, indicating that the over-expression of PTC7 enhances mitophagy induction (Figure S3). Remarkably, Kar2, a marker of endoplasmic reticulum, was not affected (Figure S4), indicating that macroautophagy was not activated by PTC7 overexpression. Taken together, these data indicate that Ptc7 regulates or participates specifically in mitophagy but not in general macroautophagy, suggesting that this specific process may compromise CLS in ptc7∆ yeast.
DISCUSSION
Our understanding of the role of mitochondria on cell physiology and metabolism has evolved in the past decades from only a bioenergetics role to an interconnected organelle whose functions exceed energy supply. A key factor for part of these mitochondrial functions is CoQ, as proper regulation of its biosynthesis pathway exceeds cell bioenergetics and is tightly linked to cellular homeostasis. This idea supports the pleiotropic effect observed in patients with primary CoQ deficiency [7][52]. Although most Coq proteins are involved in enzymatic steps required for CoQ6 biosynthesis [8], some Coq proteins play a structural function and are required to stabilize the biosynthetic complex [8][53]. In yeast, the shift from fermentation to respiration activates the expression of COQ genes to accommodate CoQ6 biosynthesis to respiratory metabolism [19][54], which is associated to regulatory mechanisms such as biosynthesis complex assembly and post-translational modifications [9][22][55][56].
–
The protein encoded by the COQ7 gene has its catalytic activity controlled by complex regulatory mechanisms [16][22][55][28][57][58]. Coq7 is a di-iron carboxylate protein with hydroxylase activity [59] that converts DMQ6 in demethyl ubiquinone. The lack of Coq7 in yeast fully abolishes CoQ6 biosynthesis and other intermediates although the expression of some point mutants such as e2519 (E223K) accumulates DMQ6 [34], which would indicate that this step is a regulatory step in this pathway. DMQ6 is also accumulated in wild type cells after the post-diauxic shift supporting this hypothesis [19][34]. A model of CoQ6 biosynthesis complex assembly, based on BN-PAGE, size exclusion chromatography and immunoprecipitation data, show that a precomplex of about 700 kDa is formed, which accumulates DMQ6 [10] that, ultimately, will be converted into CoQ6 after the recruitment of Coq7 to the complex [9][56]. Coq7 is a phosphoprotein that, in the dephosphorylated state, activates CoQ6 biosynthesis probably by interacting with the 700 kDa precomplex [22]. Here we have shown that the phosphomimetic Coq7 (Coq7-DED), which mimics Coq7 phosphorylation status in ptc7∆ strain [28], and the fully dephosphorylated Coq7 (Coq7-AAA) induced a decreased and an increased levels of CoQ6 respectively, indicating that the regulatory step on CoQ6 biosynthesis is at least partially controlled by Coq7 phosphorylation.
–
The physiological analysis of these strains confirms the catalytic function of Coq7 in CoQ6 biosynthesis and its regulatory function in mitochondrial metabolism. Expression of both Coq7-AAA and Coq7-DED decreases antioxidant protection and increases the production of endogenous oxidative stress. A similar result was reported for the ptc7∆ strain [28]. These negative effects cannot be explained by the alteration of CoQ6 levels, as the coq7∆/pmQ7 strain, which also shows high levels of CoQ6, had an endogenous oxidative stress and sensitivity similar to wild type. CoQ6 has been reported as an antioxidant molecule mainly to protect cell membranes [35][36][60] but also as a pro-oxidant agent under physio-pathological conditions [61]. CoQ is involved in ROS production in the MRC mainly because of the transfer of electrons to complex III [62][63]. It has been recently demonstrated in Drosophila that the reduced stage of CoQ (CoQH2) causes CoQH2-mediated ROS in complex I by retrograde electron transport and contributes to extend lifespan [64]. Similar conditions of higher CoQH2 in mammal-cultured cells contribute to the partial degradation of complex I by the same mechanism [65]. Thus, we propose that the production of CoQ6 in strains with low Coq7 phosphorylation are generating unbalanced CoQ6 levels and redox stages out of the MRC complexes, which does not occur in the coq7Δ/pmQ7 strain. The unbalanced levels of CoQ6 in these strains increase their sensitivity to external oxidative stress, decreasing their longevity. It has been reported previously that linolenic acid can induce mitochondrial oxidative stress [66]. The effect of linolenic acid may be enhanced by the mitochondrial dysfunction generated by the expression of pAAA and pDED versions. In fact, the expression of pAAA or pDED induces a higher level of endogenous oxidative stress and also affects the stability of respiratory complexes, which possibly make those strains more susceptible to linolenic acid-induced oxidative stress.
–
CoQ is a component of the respirasome and it is proposed that pools of CoQ are bound to specific super assembly stages of respiratory complexes [67], which depends on the carbon source [65]. Here our data show that super-complexes are dissociated in yeast strains with unbalanced CoQ6 concentrations, which also showed mitochondrial dysfunction, but not in the COQ7 multicopy transformed yeast strain (coq7∆/pmQ7) that exhibit a phenotype similar to wild type in both respiratory activities and assembly profile. Previous analyses of CoQ6 biosynthesis complex showed that Coq7 is partially located in a large size complex but it can also be detected in smaller ones even as a monomer [10]. Coq7 interacts with Coq9, which shows lipid-binding activity and Coq7 interacting domains [16]. Several steps are required to integrate the CoQ6 biosynthesis complex in yeast. First, there is an initial nucleation of Coq proteins around Coq4 [10][68] to build up the Q-synthome, which requires stabilization by Coq8 [69], an unusual protein kinase that makes the pre-complex formation [20] and demethoxy-Q6 accumulation possible [19]. At this step Coq7 is mostly phosphorylated and it is not a component of the pre-complex [10]; Coq7 must then be dephosphorylated by Ptc7 to get activated and to bind to the fully active Q-synthome [9][28][56]. We speculate that CoQ6 biosynthesis must be balanced with the components of the respiratory complexes. In light of our data, we hypothesize that the interaction of Coq7 with other proteins of the biosynthesis complex is a requirement for its integration in the MRC, and that extreme phosphorylation stages might prevent this possibility. This is supported by the data obtained from the ptc7∆ strain that mimics Coq7-DED expressing cells without eliminating all regulation of Coq7.
–
In fact, yeast contains two other mitochondrial phosphatases that belong to the same family of Ptc7, Ptc5 that participates in pyruvate dehydrogenase complex (PDH) regulation [30][70] and Ptc6/Aup1 that participates in PDH regulation and is required for mitophagy activation [31][71]. It is possible that these mitochondrial phosphatases may dephosphorylate Coq7 in the absence of Ptc7, which would explain the less severe effect on mitochondrial functions. Interestingly, the ptc7∆ strain shows shorten CLS compared to the coq7∆/pDED strain and it is not rescued by CoQ6 supplementation, although this strain shows a significant decrease of CoQ6 content. CLS is only rescued when it is transformed with the homologous gene, which is also able to rescue the CoQ6 content; overall these data indicate that Ptc7 has at least a dual function in yeast. As indicated above, Ptc6 is required for mitophagy activation [31][71] and we have shown here that Ptc7 induces mitophagy as a mechanism to recycle defective mitochondria caused by CoQ6 deficient MRC.
–
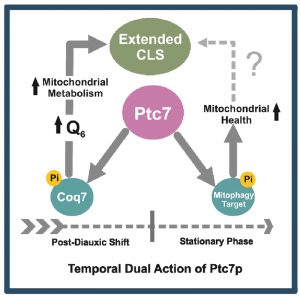
The lack of mitophagy in ptc7∆ yeast, which is a requisite to extend CLS [72][73], can explain its shorten CLS and the negative recovery after CoQ6 supplementation. We propose a dual and temporary differential function of Ptc7 on mitochondrial physiology and homeostasis (Figure 6). The entry of yeast on the post-diauxic shift (PDS) increases CoQ6 biosynthesis and therefore respiratory metabolism by Coq7 activation. However, cell homeostasis during PDS requires the recycling of the excess and defective mitochondria. Ptc7, which would trigger mitophagy by dephosphorylating an unknown target involved in this process, would be a key regulator of mitochondria homeostasis in yeast by coordinating mitochondria recycling with CoQ6 biosynthesis.
MATERIALS AND METHODS
Yeast strains and growth media
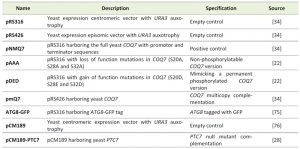
 | TABLE 1. Strains used in this study. [74] |
Yeast strains used in this study are listed in Table 1. Growth media for yeast and bacteria were prepared as described previously [22]. Yeast cells were grown at 30°C with shaking (200 rpm).
 |
Mitochondrial purification and BN-PAGE
Yeast cultures were grown in the appropriate culture media and mitochondria were purified according to the described method [74]. To solubilize mitochondria, 2 mg of pure mitochondria was incubated in 240 μl of solubilization buffer containing digitonin in a ratio 4:1 with protein, 1 mM PMSF, 10% glycerol, 150 mM potassium acetate and 30 mM HEPES, pH 7.4 for 30 min at 4°C. Solubilized samples were subjected to two rounds of centrifugation in a Beckman Coulter Microfuge 22R (15,000 × g, 15 min, at 4°C) and the supernatant was collected to BN-PAGE. Proteins quantification was performed by Bradford method (Biorad). BN-PAGE was performed with precast 3–12% gradient gels (NativePAGETM Novex® Bis-Tris Gels) using the Xcell Sure LockTM Mini-Cell electrophoresis system, including NativePAGETM anode and cathode buffers according to the company instructions at 4°C. Lanes were loaded with 200 µg of mitochondrial solubilized supernatant and as MW marker NativeMarkTM Unstained Protein Ladder was used. Gels were stained with Coomassie solution or were blotted onto PVDF, blocked in 5% Blocking Reagent (Biorad) and phosphate-buffered saline. Proteins were detected by ECL using Luminata Crescendo (Millipore) and luminescence detected by Gel Doc XR+ image processing software (Bio-Rad). Cox2p (Novex Life Technologies), porin (Invitrogen) and Coq7 (Gift of Dr. C. F. Clarke, UCLA, USA) primary antibodies were used at 1: 2,000, 1: 1,000 and 1: 2,000 respectively. Goat anti-mouse and anti-rabbit secondary antibodies conjugated to horseradish peroxidase (Calbiochem) were used at a 1:5,000 dilutions.
–
Mitochondrial respiratory chain (MRC) activities
Fresh mitochondria were used to measure NADH-cytochrome c reductase, succinate-cytochrome c reductase activities and superoxide generation. Other MRC activities (NADH-DCIP reductase, succinate-DCIP reductase and decylubiquinol-cytochrome c reductase), were performed with fresh samples subjected to one freeze-thaw cycle. All MRC activities were determined according to previously published methods [34]. Superoxide generation was measured using the same method than for measuring complex III activity (decylubiquinol-cytochrome c reductase) but using acetylated cytochrome c instead cytochrome c [33]. H2O2 generation was performed using Amplex Red kit (Invitrogen) according to the manufacturer instructions.
–
Chronological Life Span (CLS)
Analysis was performed as previously described [37]. Briefly, cells were incubated in YPD and CLS was monitored starting at day 3 by quantification of colony forming units (CFUs) every 48 hours using the software OpenCFU 3.9 beta. The number of CFUs at day three was considered as 100% survival. Survival log-rank analyses Sigmastat 3.0 (SPSS) were calculated for each pair of lifespan analyses and average lifespan were shown in the corresponding dataset in Supplementary Material.
–
Protein mass spectrometry identification
Acrylamide gel bands were distained in NH4HCO3 25 mM water/ACN 50:50. For reduction of cysteines, samples were incubated at 56°C for 60 min in 10 mM DTT (NH4HCO3 25 mM). Cysteine carbamidomethylation was performed embedding bands in 55 mM IAA solution and incubating at room temperature for 30 min. Samples were in gel digested by trypsin (0.2 µg/µl in 1 mM HCl) diluted in NH4HCO3. Gel bands were covered with enzyme solution and incubated at 30°C overnight. Reaction was stopped with acetonitrile, and peptide were extracted adding 0.2% TFA. Prior to protein identification by MALDI-MS, we used nano-liquid chromatography for reversed phase peptide separation. Peptide fractioning was performed in a Proxeon EASY-nLC II apparatus with a C18 column (EASY-column, 75 µm x 100 mm) and mobile phases: Buffer A: 0.1% TFA (H2O) and Buffer B: 0.1% TFA (ACN). Elution process was divided in the following flow steps: 0-48 min gradient 2%-45% B; 48-50 min gradient 45-100% B; 50-60 min isocratic 100% B. Fractions were collected every 15” using a Bruker Proteineer fc fraction collector and spotted onto a MALDI target plate. 192 samples were spotted and overlaid with 0.5 µl drops of HCCA matrix solution and left air dry. MALDI measurements were performed in a Bruker Ultraflextreme MALDI-TOF/TOF system, using Bruker Peptide Calibration standards as mass standards. A MALDI fingerprint spectrum was obtained for each fraction and peaks with higher intensity in each spot were selected as mass precursors for MS/MS peptide fragmentation experiments. Bruker WARP-LC software was used to process spectra and to integrate chromatography fractions data. Protein identification was carried out using MASCOT server. Ammonium bicarbonate, DL-dithiothreitol (DTT), Iodoacetamide (IAA), trypsin from porcine pancreas, trifluoroacetic acid (TFA) and α-cyano-hydroxycinnamc acid (HCCA) were purchased from Sigma-Aldrich. Water (HPLC grade) and acetonitrile (HPLC grade) were purchased from Fluka. Peptide calibration standards were purchased from Bruker.
–
Total yeast protein extraction
Cells (10.106 in 500 µl of water) were disrupted with 100 µl 2 M NaOH and 35% β-mercaptoethanol for 15 min on ice. Proteins were precipitated after the addition of 100 µl 3 M TCA for 15 min on ice. The pellet obtained after 15 min centrifugation on a microcentrifuge at full speed was washed with acetone, dried and resuspended in 30 µl of SDS-PAGE 1 x LB.
–
Other methods
CoQ6 quantification was performed using mitochondrial samples according to previously published methods [28]. Densitometry analysis was carried out with a Gel Doc XR+ (Bio-Rad) with Image Lab 4.0 as software analysis. Statistical (t-Student) analyses were carried out using the Sigmastat 3.0 (SPSS) statistical package. Mitochondrial DNA integrity was checked in all strains using two ρ0 strains, JM6 and JM8 strains. All results are expressed as the average ± SD. Statistical analyses were carried out using the Sigmastat 3.0 (SPSS) statistical package.
References
- M. Doimo, M.A. Desbats, C. Cerqua, M. Cassina, E. Trevisson, and L. Salviati, "Genetics of Coenzyme Q10 Deficiency", Molecular Syndromology, vol. 5, pp. 156-162, 2014. http://dx.doi.org/10.1159/000362826
- V. Emmanuele, L.C. López, A. Berardo, A. Naini, S. Tadesse, B. Wen, E. D’Agostino, M. Solomon, S. DiMauro, C. Quinzii, and M. Hirano, "Heterogeneity of Coenzyme Q10Deficiency", Archives of Neurology, vol. 69, 2012. http://dx.doi.org/10.1001/archneurol.2012.206
- S. DiMauro, C.M. Quinzii, and M. Hirano, "Mutations in coenzyme Q10 biosynthetic genes", Journal of Clinical Investigation, vol. 117, pp. 587-589, 2007. http://dx.doi.org/10.1172/JCI31423
- M. Peng, M.J. Falk, V.H. Haase, R. King, E. Polyak, M. Selak, M. Yudkoff, W.W. Hancock, R. Meade, R. Saiki, A.L. Lunceford, C.F. Clarke, and D. L. Gasser, "Primary Coenzyme Q Deficiency in Pdss2 Mutant Mice Causes Isolated Renal Disease", PLoS Genetics, vol. 4, pp. e1000061, 2008. http://dx.doi.org/10.1371/journal.pgen.1000061
- C.M. Quinzii, and M. Hirano, "Primary and secondary CoQ10 deficiencies in humans", BioFactors, vol. 37, pp. 361-365, 2011. http://dx.doi.org/10.1002/biof.155
- D. Yubero, R. Montero, M.A. Martín, J. Montoya, A. Ribes, M. Grazina, E. Trevisson, J.C. Rodriguez-Aguilera, I.P. Hargreaves, L. Salviati, P. Navas, R. Artuch, C. Jou, C. Jimenez-Mallebrera, A. Nascimento, B. Pérez-Dueñas, C. Ortez, F. Ramos, J. Colomer, M. O’Callaghan, M. Pineda, A. García-Cazorla, C. Espinós, A. Ruiz, A. Macaya, A. Marcé-Grau, J. Garcia-Villoria, A. Arias, S. Emperador, E. Ruiz-Pesini, E. Lopez-Gallardo, V. Neergheen, M. Simões, L. Diogo, A. Blázquez, A. González-Quintana, A. Delmiro, C. Domínguez-González, J. Arenas, M.T. García-Silva, E. Martín, P. Quijada, A. Hernández-Laín, M. Morán, E. Rivas Infante, R. Ávila Polo, C. Paradas Lópe, J. Bautista Lorite, E.M. Martínez Fernández, A.B. Cortés, A. Sánchez-Cuesta, M.V. Cascajo, M. Alcázar, and G. Brea-Calvo, "Secondary coenzyme Q 10 deficiencies in oxidative phosphorylation (OXPHOS) and non-OXPHOS disorders", Mitochondrion, vol. 30, pp. 51-58, 2016. http://dx.doi.org/10.1016/j.mito.2016.06.007
- C. Santos-Ocaña, L. Salviati, and P. Navas, "The genes of CoQ10. In: Iain P. Hargreaves, Ph.D. (Neurometabolic Unit, National Hospital, Queen Square, London, UK) Assistant Editor: April K. Hargreaves, Ph.D. (Trinity University, Dublin I (ed) Coenzyme Q10 From fact to Fict., 1st ed", Nova Science Publisher, New York; pp 205–226, 2015.
- U.C. Tran, and C.F. Clarke, "Endogenous synthesis of coenzyme Q in eukaryotes", Mitochondrion, vol. 7, pp. S62-S71, 2007. http://dx.doi.org/10.1016/j.mito.2007.03.007
- I. González‐Mariscal, E. García‐Testón, S. Padilla, A. Martín‐Montalvo, T. Pomares‐Viciana, L. Vazquez‐Fonseca, P. Gandolfo‐Domínguez, and C. Santos‐Ocaña, "Regulation of coenzyme Q biosynthesis in yeast: A new complex in the block", IUBMB Life, vol. 66, pp. 63-70, 2014. http://dx.doi.org/10.1002/iub.1243
- B. Marbois, P. Gin, M. Gulmezian, and C.F. Clarke, "The yeast Coq4 polypeptide organizes a mitochondrial protein complex essential for coenzyme Q biosynthesis", Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids, vol. 1791, pp. 69-75, 2009. http://dx.doi.org/10.1016/j.bbalip.2008.10.006
- C.J. Leonard, L. Aravind, and E.V. Koonin, "Novel families of putative protein kinases in bacteria and archaea: evolution of the "eukaryotic" protein kinase superfamily.", Genome research, 1998. http://www.ncbi.nlm.nih.gov/pubmed/9799791
- D.R. Macinga, G.M. Cook, R.K. Poole, and P.N. Rather, "Identification and characterization of aarF, a locus required for production of ubiquinone in Providencia stuartii and Escherichia coli and for expression of 2'-N-acetyltransferase in P. stuartii.", Journal of bacteriology, 1998. http://www.ncbi.nlm.nih.gov/pubmed/9422602
- J. Stefely, A. Reidenbach, A. Ulbrich, K. Oruganty, B. Floyd, A. Jochem, J. Saunders, I. Johnson, C. Minogue, R. Wrobel, G. Barber, D. Lee, S. Li, N. Kannan, J. Coon, C. Bingman, and D. Pagliarini, "Mitochondrial ADCK3 Employs an Atypical Protein Kinase-like Fold to Enable Coenzyme Q Biosynthesis", Molecular Cell, vol. 57, pp. 83-94, 2015. http://dx.doi.org/10.1016/j.molcel.2014.11.002
- J. Stefely, F. Licitra, L. Laredj, A. Reidenbach, Z. Kemmerer, A. Grangeray, T. Jaeg-Ehret, C. Minogue, A. Ulbrich, P. Hutchins, E. Wilkerson, Z. Ruan, D. Aydin, A. Hebert, X. Guo, E. Freiberger, L. Reutenauer, A. Jochem, M. Chergova, I. Johnson, D. Lohman, M. Rush, N. Kwiecien, P. Singh, A. Schlagowski, B. Floyd, U. Forsman, P. Sindelar, M. Westphall, F. Pierrel, J. Zoll, M. Dal Peraro, N. Kannan, C. Bingman, J. Coon, P. Isope, H. Puccio, and D. Pagliarini, "Cerebellar Ataxia and Coenzyme Q Deficiency through Loss of Unorthodox Kinase Activity", Molecular Cell, vol. 63, pp. 608-620, 2016. http://dx.doi.org/10.1016/j.molcel.2016.06.030
- E.J. Hsieh, P. Gin, M. Gulmezian, U.C. Tran, R. Saiki, B.N. Marbois, and C.F. Clarke, "Saccharomyces cerevisiae Coq9 polypeptide is a subunit of the mitochondrial coenzyme Q biosynthetic complex", Archives of Biochemistry and Biophysics, vol. 463, pp. 19-26, 2007. http://dx.doi.org/10.1016/j.abb.2007.02.016
- D.C. Lohman, F. Forouhar, E.T. Beebe, M.S. Stefely, C.E. Minogue, A. Ulbrich, J.A. Stefely, S. Sukumar, M. Luna-Sánchez, A. Jochem, S. Lew, J. Seetharaman, R. Xiao, H. Wang, M.S. Westphall, R.L. Wrobel, J.K. Everett, J.C. Mitchell, L.C. López, J.J. Coon, L. Tong, and D.J. Pagliarini, "Mitochondrial COQ9 is a lipid-binding protein that associates with COQ7 to enable coenzyme Q biosynthesis", Proceedings of the National Academy of Sciences, vol. 111, 2014. http://dx.doi.org/10.1073/pnas.1413128111
- P. Gin, and C.F. Clarke, "Genetic Evidence for a Multi-subunit Complex in Coenzyme Q Biosynthesis in Yeast and the Role of the Coq1 Hexaprenyl Diphosphate Synthase", Journal of Biological Chemistry, vol. 280, pp. 2676-2681, 2005. http://dx.doi.org/10.1074/jbc.M411527200
- B. Marbois, P. Gin, K.F. Faull, W.W. Poon, P.T. Lee, J. Strahan, J.N. Shepherd, and C.F. Clarke, "Coq3 and Coq4 Define a Polypeptide Complex in Yeast Mitochondria for the Biosynthesis of Coenzyme Q", Journal of Biological Chemistry, vol. 280, pp. 20231-20238, 2005. http://dx.doi.org/10.1074/jbc.M501315200
- S. Padilla, U.C. Tran, M. Jiménez-Hidalgo, J.M. López-Martín, A. Martín-Montalvo, C.F. Clarke, P. Navas, and C. Santos-Ocaña, "Hydroxylation of demethoxy-Q6 constitutes a control point in yeast coenzyme Q6 biosynthesis", Cellular and Molecular Life Sciences, vol. 66, pp. 173-186, 2008. http://dx.doi.org/10.1007/s00018-008-8547-7
- C.H. He, L.X. Xie, C.M. Allan, U.C. Tran, and C.F. Clarke, "Coenzyme Q supplementation or over-expression of the yeast Coq8 putative kinase stabilizes multi-subunit Coq polypeptide complexes in yeast coq null mutants", Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids, vol. 1841, pp. 630-644, 2014. http://dx.doi.org/10.1016/j.bbalip.2013.12.017
- L.X. Xie, M. Ozeir, J.Y. Tang, J.Y. Chen, S. Jaquinod, M. Fontecave, C.F. Clarke, and F. Pierrel, "Overexpression of the Coq8 Kinase in Saccharomyces cerevisiae coq Null Mutants Allows for Accumulation of Diagnostic Intermediates of the Coenzyme Q6 Biosynthetic Pathway", Journal of Biological Chemistry, vol. 287, pp. 23571-23581, 2012. http://dx.doi.org/10.1074/jbc.M112.360354
- A. Martín-Montalvo, I. González-Mariscal, S. Padilla, M. Ballesteros, D.L. Brautigan, P. Navas, and C. Santos-Ocaña, "Respiratory-induced coenzyme Q biosynthesis is regulated by a phosphorylation cycle of Cat5p/Coq7p", Biochemical Journal, vol. 440, pp. 107-114, 2011. http://dx.doi.org/10.1042/BJ20101422
- B.N. Marbois, and C.F. Clarke, "The COQ7 Gene Encodes a Protein in Saccharomyces cerevisiae Necessary for Ubiquinone Biosynthesis", Journal of Biological Chemistry, vol. 271, pp. 2995-3004, 1996. http://dx.doi.org/10.1074/jbc.271.6.2995
- A. Tauche, U. Krause-Buchholz, and G. Rödel, "Ubiquinone biosynthesis inSaccharomyces cerevisiae: the molecular organization ofO-methylase Coq3p depends on Abc1p/Coq8p", FEMS Yeast Research, vol. 8, pp. 1263-1275, 2008. http://dx.doi.org/10.1111/j.1567-1364.2008.00436.x
- L.X. Xie, E.J. Hsieh, S. Watanabe, C.M. Allan, J.Y. Chen, U.C. Tran, and C.F. Clarke, "Expression of the human atypical kinase ADCK3 rescues coenzyme Q biosynthesis and phosphorylation of Coq polypeptides in yeast coq8 mutants", Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids, vol. 1811, pp. 348-360, 2011. http://dx.doi.org/10.1016/j.bbalip.2011.01.009
- C. Busso, J.R. Ferreira-Júnior, J.A. Paulela, L. Bleicher, M. Demasi, and M.H. Barros, "Coq7p relevant residues for protein activity and stability", Biochimie, vol. 119, pp. 92-102, 2015. http://dx.doi.org/10.1016/j.biochi.2015.10.016
- D. Barford, A.K. Das, and M. Egloff, "THE STRUCTURE AND MECHANISM OF PROTEIN PHOSPHATASES: Insights into Catalysis and Regulation", Annual Review of Biophysics and Biomolecular Structure, vol. 27, pp. 133-164, 1998. http://dx.doi.org/10.1146/annurev.biophys.27.1.133
- A. Martín-Montalvo, I. González-Mariscal, T. Pomares-Viciana, S. Padilla-López, M. Ballesteros, L. Vazquez-Fonseca, P. Gandolfo, D.L. Brautigan, P. Navas, and C. Santos-Ocaña, "The Phosphatase Ptc7 Induces Coenzyme Q Biosynthesis by Activating the Hydroxylase Coq7 in Yeast", Journal of Biological Chemistry, vol. 288, pp. 28126-28137, 2013. http://dx.doi.org/10.1074/jbc.M113.474494
- U. Krause-Buchholz, U. Gey, J. Wünschmann, S. Becker, and G. Rödel, "YIL042c and YOR090c encode the kinase and phosphatase of the Saccharomyces cerevisiae pyruvate dehydrogenase complex", FEBS Letters, vol. 580, pp. 2553-2560, 2006. http://dx.doi.org/10.1016/j.febslet.2006.04.002
- U. Gey, C. Czupalla, B. Hoflack, G. Rödel, and U. Krause-Buchholz, "Yeast Pyruvate Dehydrogenase Complex Is Regulated by a Concerted Activity of Two Kinases and Two Phosphatases", Journal of Biological Chemistry, vol. 283, pp. 9759-9767, 2008. http://dx.doi.org/10.1074/jbc.M708779200
- R. Tal, G. Winter, N. Ecker, D.J. Klionsky, and H. Abeliovich, "Aup1p, a Yeast Mitochondrial Protein Phosphatase Homolog, Is Required for Efficient Stationary Phase Mitophagy and Cell Survival", Journal of Biological Chemistry, vol. 282, pp. 5617-5624, 2007. http://dx.doi.org/10.1074/jbc.m605940200
- H. Ruan, Z. Yan, H. Sun, and L. Jiang, "TheYCR079wgene confers a rapamycin-resistant function and encodes the sixth type 2C protein phosphatase inSaccharomyces cerevisiae", FEMS Yeast Research, vol. 7, pp. 209-215, 2007. http://dx.doi.org/10.1111/j.1567-1364.2006.00160.x
- C. Santos-Ocaña, J.M. Villalba, F. Córdoba, S. Padilla, F.L. Crane, C.F. Clarke, and P. Navas, "Genetic evidence for coenzyme Q requirement in plasma membrane electron transport.", Journal of bioenergetics and biomembranes, 1998. http://www.ncbi.nlm.nih.gov/pubmed/9932649
- S. Padilla, T. Jonassen, M.A. Jiménez-Hidalgo, D.J.M. Fernández-Ayala, G. López-Lluch, B. Marbois, P. Navas, C.F. Clarke, and C. Santos-Ocaña, "Demethoxy-Q, An Intermediate of Coenzyme Q Biosynthesis, Fails to Support Respiration in Saccharomyces cerevisiae and Lacks Antioxidant Activity", Journal of Biological Chemistry, vol. 279, pp. 25995-26004, 2004. http://dx.doi.org/10.1074/jbc.M400001200
- W.W. Poon, T.Q. Do, B. Noelle Marbois, and C.F. Clarke, "Sensitivity to treatment with polyunsaturated fatty acids is a general characteristic of the ubiquinone-deficient yeast coq mutants", Molecular Aspects of Medicine, vol. 18, pp. 121-127, 1997. http://dx.doi.org/10.1016/S0098-2997(97)00004-6
- T.Q. Do, J.R. Schultz, and C.F. Clarke, "Enhanced sensitivity of ubiquinone-deficient mutants of Saccharomyces cerevisiae to products of autoxidized polyunsaturated fatty acids.", Proceedings of the National Academy of Sciences, vol. 93, pp. 7534-7539, 1996. http://dx.doi.org/10.1073/pnas.93.15.7534
- P. Fabrizio, and V.D. Longo, "The chronological life span of Saccharomyces cerevisiae.", Aging cell, 2003. http://www.ncbi.nlm.nih.gov/pubmed/12882320
- M.H. Barros, B. Bandy, E.B. Tahara, and A.J. Kowaltowski, "Higher Respiratory Activity Decreases Mitochondrial Reactive Oxygen Release and Increases Life Span in Saccharomyces cerevisiae", Journal of Biological Chemistry, vol. 279, pp. 49883-49888, 2004. http://dx.doi.org/10.1074/jbc.M408918200
- A.M. Aerts, P. Zabrocki, G. Govaert, J. Mathys, D. Carmona-Gutierrez, F. Madeo, J. Winderickx, B.P. Cammue, and K. Thevissen, "Mitochondrial dysfunction leads to reduced chronological lifespan and increased apoptosis in yeast", FEBS Letters, vol. 583, pp. 113-117, 2008. http://dx.doi.org/10.1016/j.febslet.2008.11.028
- M. Breitenbach, P. Laun, J.R. Dickinson, A. Klocker, M. Rinnerthaler, I.W. Dawes, M.T. Aung-Htut, L. Breitenbach-Koller, A. Caballero, T. Nyström, S. Büttner, T. Eisenberg, F. Madeo, and M. Ralser, "The Role of Mitochondria in the Aging Processes of Yeast", Subcellular Biochemistry, pp. 55-78, 2011. http://dx.doi.org/10.1007/978-94-007-2561-4_3
- A. Ocampo, J. Liu, E. Schroeder, G. Shadel, and A. Barrientos, "Mitochondrial Respiratory Thresholds Regulate Yeast Chronological Life Span and its Extension by Caloric Restriction", Cell Metabolism, vol. 16, pp. 55-67, 2012. http://dx.doi.org/10.1016/j.cmet.2012.05.013
- V.D. Longo, L. Liou, J.S. Valentine, and E.B. Gralla, "Mitochondrial Superoxide Decreases Yeast Survival in Stationary Phase", Archives of Biochemistry and Biophysics, vol. 365, pp. 131-142, 1999. http://dx.doi.org/10.1006/abbi.1999.1158
- P. Fabrizio, L. Liou, V.N. Moy, A. Diaspro, J.S. Valentine, E.B. Gralla, and V.D. Longo, "SOD2 functions downstream of Sch9 to extend longevity in yeast.", Genetics, 2003. http://www.ncbi.nlm.nih.gov/pubmed/12586694
- S. Gonidakis, and V.D. Longo, "Oxidative Stress and Aging in the Budding Yeast Saccharomyces cerevisiae", Oxidative Stress in Aging, pp. 67-79, 2008. http://dx.doi.org/10.1007/978-1-59745-420-9_5
- C. Santos-Ocaña, T.Q. Do, C.F. Clarke, S. Padilla, and P. Navas, "Uptake of Exogenous Coenzyme Q and Transport to Mitochondria Is Required for bc1 Complex Stability in Yeast coq Mutants", Journal of Biological Chemistry, vol. 277, pp. 10973-10981, 2002. http://dx.doi.org/10.1074/jbc.M112222200
- H. Schagger, "Supercomplexes in the respiratory chains of yeast and mammalian mitochondria", The EMBO Journal, vol. 19, pp. 1777-1783, 2000. http://dx.doi.org/10.1093/emboj/19.8.1777
- R.J. Youle, and D.P. Narendra, "Mechanisms of mitophagy", Nature Reviews Molecular Cell Biology, vol. 12, pp. 9-14, 2010. http://dx.doi.org/10.1038/nrm3028
- V.R. Richard, A. Leonov, A. Beach, M.T. Burstein, O. Koupaki, A. Gomez-Perez, S. Levy, L. Pluska, S. Mattie, R. Rafeh, T. Iouk, S. Sheibani, M. Greenwood, H. Vali, and V.I. Titorenko, "Macromitophagy is a longevity assurance process that in chronologically aging yeast limited in calorie supply sustains functional mitochondria and maintains cellular lipid homeostasis", Aging, vol. 5, pp. 234-269, 2013. http://dx.doi.org/10.18632/aging.100547
- �. Rodríguez-Hernández, M.D. Cordero, L. Salviati, R. Artuch, M. Pineda, P. Briones, L. Gómez Izquierdo, D. Cotán, P. Navas, and J.A. Sánchez-Alcázar, "Coenzyme Q deficiency triggers mitochondria degradation by mitophagy", Autophagy, vol. 5, pp. 19-32, 2009. http://dx.doi.org/10.4161/auto.5.1.7174
- T. Shintani, and D.J. Klionsky, "Autophagy in Health and Disease: A Double-Edged Sword", Science, vol. 306, pp. 990-995, 2004. http://dx.doi.org/10.1126/science.1099993
- I. Kissová, M. Deffieu, S. Manon, and N. Camougrand, "Uth1p Is Involved in the Autophagic Degradation of Mitochondria", Journal of Biological Chemistry, vol. 279, pp. 39068-39074, 2004. http://dx.doi.org/10.1074/jbc.M406960200
- L.N. Laredj, F. Licitra, and H.M. Puccio, "The molecular genetics of coenzyme Q biosynthesis in health and disease", Biochimie, vol. 100, pp. 78-87, 2014. http://dx.doi.org/10.1016/j.biochi.2013.12.006
- M. Kawamukai, "Biosynthesis of coenzyme Q in eukaryotes", Bioscience, Biotechnology, and Biochemistry, vol. 80, pp. 23-33, 2016. http://dx.doi.org/10.1080/09168451.2015.1065172
- R.A. Hagerman, and R.A. Willis, "The yeast gene COQ5 is differentially regulated by Mig1p, Rtg3p and Hap2p", Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression, vol. 1578, pp. 51-58, 2002. http://dx.doi.org/10.1016/S0167-4781(02)00496-7
- C.M. Allan, A.M. Awad, J.S. Johnson, D.I. Shirasaki, C. Wang, C.E. Blaby-Haas, S.S. Merchant, J.A. Loo, and C.F. Clarke, "Identification of Coq11, a New Coenzyme Q Biosynthetic Protein in the CoQ-Synthome in Saccharomyces cerevisiae", Journal of Biological Chemistry, vol. 290, pp. 7517-7534, 2015. http://dx.doi.org/10.1074/jbc.M114.633131
- G. Brea-Calvo, E. Siendones, J.A. Sánchez-Alcázar, R. de Cabo, and P. Navas, "Cell Survival from Chemotherapy Depends on NF-κB Transcriptional Up-Regulation of Coenzyme Q Biosynthesis", PLoS ONE, vol. 4, pp. e5301, 2009. http://dx.doi.org/10.1371/journal.pone.0005301
- M.V. Cascajo, K. Abdelmohsen, J.H. Noh, D.J. Fernández-Ayala, I.M. Willers, G. Brea, G. López-Lluch, M. Valenzuela-Villatoro, J.M. Cuezva, M. Gorospe, E. Siendones, and P. Navas, "RNA-binding proteins regulate cell respiration and coenzyme Q biosynthesis by post-transcriptional regulation ofCOQ7", RNA Biology, vol. 13, pp. 622-634, 2016. http://dx.doi.org/10.1080/15476286.2015.1119366
- P. Stenmark, J. Grünler, J. Mattsson, P.J. Sindelar, P. Nordlund, and D.A. Berthold, "A New Member of the Family of Di-iron Carboxylate Proteins", Journal of Biological Chemistry, vol. 276, pp. 33297-33300, 2001. http://dx.doi.org/10.1074/jbc.C100346200
- A. MAROZ, R. ANDERSON, R. SMITH, and M. MURPHY, "Reactivity of ubiquinone and ubiquinol with superoxide and the hydroperoxyl radical: implications for in vivo antioxidant activity", Free Radical Biology and Medicine, vol. 46, pp. 105-109, 2009. http://dx.doi.org/10.1016/j.freeradbiomed.2008.09.033
- H. Nohl, L. Gille, K. SchÖnheit, and Y. Liu, "Conditions allowing redox-cycling ubisemiquinone in mitochondria to establish a direct redox couple with molecular oxygen", Free Radical Biology and Medicine, vol. 20, pp. 207-213, 1996. http://dx.doi.org/10.1016/0891-5849(95)02038-1
- A. Boveris, E. Cadenas, and A.O.M. Stoppani, "Role of ubiquinone in the mitochondrial generation of hydrogen peroxide", Biochemical Journal, vol. 156, pp. 435-444, 1976. http://dx.doi.org/10.1042/bj1560435
- J.F. Turrens, A. Alexandre, and A.L. Lehninger, "Ubisemiquinone is the electron donor for superoxide formation by complex III of heart mitochondria", Archives of Biochemistry and Biophysics, vol. 237, pp. 408-414, 1985. http://dx.doi.org/10.1016/0003-9861(85)90293-0
- F. Scialo, V. Mallikarjun, R. Stefanatos, and A. Sanz, "Regulation of Lifespan by the Mitochondrial Electron Transport Chain: Reactive Oxygen Species-Dependent and Reactive Oxygen Species-Independent Mechanisms", Antioxidants & Redox Signaling, vol. 19, pp. 1953-1969, 2013. http://dx.doi.org/10.1089/ars.2012.4900
- A. Guarás, E. Perales-Clemente, E. Calvo, R. Acín-Pérez, M. Loureiro-Lopez, C. Pujol, I. Martínez-Carrascoso, E. Nuñez, F. García-Marqués, M.A. Rodríguez-Hernández, A. Cortés, F. Diaz, A. Pérez-Martos, C.T. Moraes, P. Fernández-Silva, A. Trifunovic, P. Navas, J. Vazquez, and J.A. Enríquez, "The CoQH2/CoQ Ratio Serves as a Sensor of Respiratory Chain Efficiency", Cell Reports, vol. 15, pp. 197-209, 2016. http://dx.doi.org/10.1016/j.celrep.2016.03.009
- A. Colquhoun, and R.I. Schumacher, "γ-Linolenic acid and eicosapentaenoic acid induce modifications in mitochondrial metabolism, reactive oxygen species generation, lipid peroxidation and apoptosis in Walker 256 rat carcinosarcoma cells", Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids, vol. 1533, pp. 207-219, 2001. http://dx.doi.org/10.1016/s1388-1981(01)00136-6
- E. Lapuente-Brun, R. Moreno-Loshuertos, R. Acín-Pérez, A. Latorre-Pellicer, C. Colás, E. Balsa, E. Perales-Clemente, P.M. Quirós, E. Calvo, M.A. Rodríguez-Hernández, P. Navas, R. Cruz, �. Carracedo, C. López-Otín, A. Pérez-Martos, P. Fernández-Silva, E. Fernández-Vizarra, and J.A. Enríquez, "Supercomplex Assembly Determines Electron Flux in the Mitochondrial Electron Transport Chain", Science, vol. 340, pp. 1567-1570, 2013. http://dx.doi.org/10.1126/science.1230381
- I. González-Mariscal, E. García-Testón, S. Padilla, A. Martín-Montalvo, T. Pomares Viciana, L. Vazquez-Fonseca, P. Gandolfo Domínguez, and C. Santos-Ocaña, "The Regulation of Coenzyme Q Biosynthesis in Eukaryotic Cells: All That Yeast Can Tell Us", Molecular Syndromology, vol. 5, pp. 107-118, 2014. http://dx.doi.org/10.1159/000362897
- T.Q. Do, A.Y. Hsu, T. Jonassen, P.T. Lee, and C.F. Clarke, "A Defect in Coenzyme Q Biosynthesis Is Responsible for the Respiratory Deficiency in Saccharomyces cerevisiae abc1Mutants", Journal of Biological Chemistry, vol. 276, pp. 18161-18168, 2001. http://dx.doi.org/10.1074/jbc.M100952200
- J. Ariño, A. Casamayor, and A. González, "Type 2C Protein Phosphatases in Fungi", Eukaryotic Cell, vol. 10, pp. 21-33, 2011. http://dx.doi.org/10.1128/EC.00249-10
- D. Journo, A. Mor, and H. Abeliovich, "Aup1-mediated Regulation of Rtg3 during Mitophagy", Journal of Biological Chemistry, vol. 284, pp. 35885-35895, 2009. http://dx.doi.org/10.1074/jbc.M109.048140
- H. Abeliovich, "Stationary-Phase Mitophagy in RespiringSaccharomyces cerevisiae", Antioxidants & Redox Signaling, vol. 14, pp. 2003-2011, 2011. http://dx.doi.org/10.1089/ars.2010.3807
- . , B. Sampaio-Marques, W. Burhans, P. Ludovico, . , and . , "Longevity pathways and maintenance of the proteome: the role of autophagy and mitophagy during yeast ageing", Microbial Cell, vol. 1, pp. 118-127, 2014. http://dx.doi.org/10.15698/mic2014.04.136
- B.S. Glick, and L.A. Pon, "[14] Isolation of highly purified mitochondria from Saccharomyces cerevisiae", Methods in Enzymology, pp. 213-223, 1995. http://dx.doi.org/10.1016/0076-6879(95)60139-2
SUPPLEMENTAL INFORMATION
![]() Download Supplemental Information
Download Supplemental Information
ACKNOWLEDGMENTS
The research group is funded by the Andalusian Government as the BIO177 group through FEDER funds (European Commission), by the Ministerio de Economía y Competitividad, Instituto de Salud Carlos III (FIS PI14/01962) and by the International Q10 Association.
AMMS received a predoctoral fellowship from the Consejería de Innovación Ciencia y Empresa, Junta de Andalucía (Spain). IGM received a predoctoral fellowship from the Plan Propio of the Universidad Pablo de Olavide de Sevilla.
The authors thank the group components for the critical reading of the manuscript and also to Ana Sanchez Cuesta for her technical help.
COPYRIGHT
© 2017

Balanced CoQ6 biosynthesis is required for lifespan and mitophagy in yeast by I. González-Mariscal et al is licensed under a Creative Commons Attribution 4.0 International License.