Back to article: Evolution of substrate specificity in the Nucleobase-Ascorbate Transporter (NAT) protein family
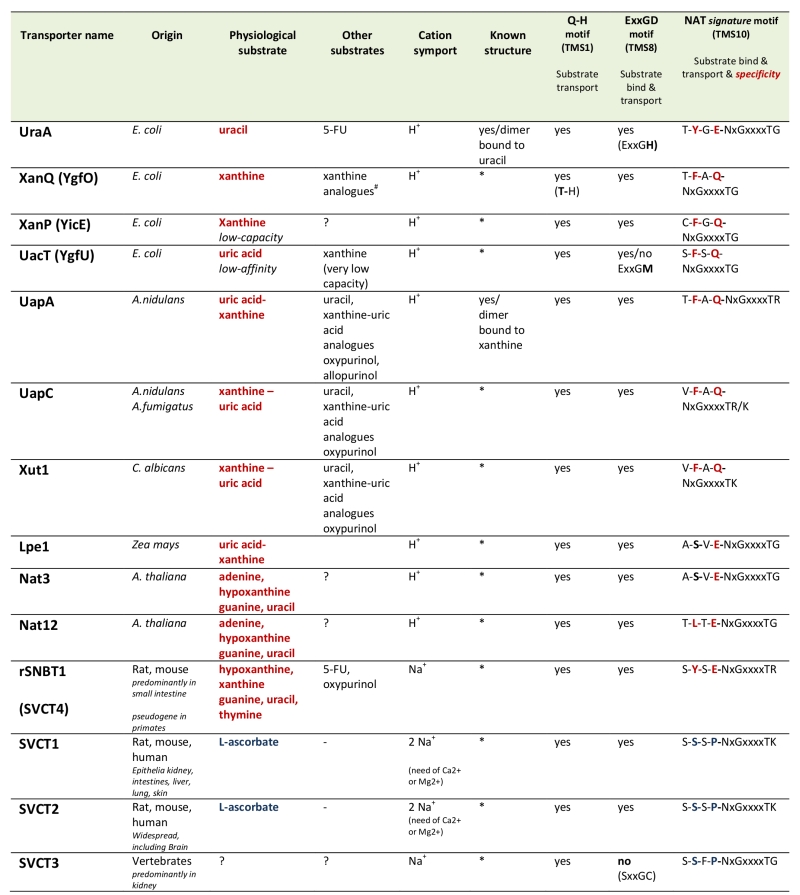
Table 1. Characteristics of biochemically characterized NATs.
All NATs shown are high-affinity transporters recognizing their physiological substrate at the low μΜ range, except UacT which is low affinity transporter. NATs marked with an (*) can be modeled, with 100% probability, on the crystal structure of UapA or UraA conforming to the 7+7 inverted repeat fold [35]. 5-FU is 5-fluorouracil. Consensus sequences of three motifs involved in function and specificity are shown. Notice that the most significant differences in the NAT signature motif of nucleobase versus L-ascorbate transporters concern substitutions of the aromatic Phe/Tyr at position 2 and of the polar Gln/Glu with a Pro. ?, not tested. #S. Frillingos pers. com.
35. Västermark A, and Saier MH Jr (2014). Evolutionary relationship between 5+5 and 7+7 inverted repeat folds within the amino acid-polyamine-organocation superfamily. Proteins 82(2):336-346. http://dx.doi.org/10.1002/prot.24401