Research Reports:
Microbial Cell, Vol. 1, No. 7, pp. 241 - 246; doi: 10.15698/mic2014.07.154
Exogenous addition of histidine reduces copper availability in the yeast Saccharomyces cerevisiae
Graduate School of Biological Sciences, Nara Institute of Science and Technology, 8916-5 Takayama, Ikoma, Nara 630-0192, Japan.
Keywords: yeast Saccharomyces cerevisiae, basic amino acids, histidine cytotoxicity, copper transporter Ctr1, mitochondrial respiration.
Received originally: 04/04/2014 Received in revised form: 09/06/2014
Accepted: 19/06/2014
Published: 07/07/2014
Correspondence:
Hiroshi Takagi, Graduate School of Biological Sciences, Nara Institute of Science and Technology, 8916-5 Takayama, Ikoma; Nara 630-0192, Japan hiro@bs.naist.jp
Conflict of interest statement: The authors declare no conflict of interest.
Please cite this article as: Daisuke Watanabe, Rie Kikushima, Miho Aitoku, Akira Nishimura, Iwao Ohtsu, Ryo Nasuno, and Hiroshi Takagi (2014). Exogenous addition of histidine reduces copper availability in the yeast Saccharomyces cerevisiae. Microbial Cell 1(7): 241-246.
Abstract
The basic amino acid histidine inhibited yeast cell growth more severely than lysine and arginine. Overexpression of CTR1, which encodes a high-affinity copper transporter on the plasma membrane, or addition of copper to the medium alleviated this cytotoxicity. However, the intracellular level of copper ions was not decreased in the presence of excess histidine. These results indicate that histidine cytotoxicity is associated with low copper availability inside cells, not with impaired copper uptake. Furthermore, histidine did not affect cell growth under limited respiration conditions, suggesting that histidine cytotoxicity is involved in deficiency of mitochondrial copper.
INTRODUCTION
Free amino acids play pivotal roles as building blocks of proteins, as intermediates in metabolism, and also as regulators of a wide variety of cellular functions. In our previous studies, several amino acids including proline and arginine show cryoprotective activity in the budding yeast Saccharomyces cerevisiae [1][2][3]. We recently showed that, under oxidative stress conditions, increased conversion of proline into arginine led to the flavoprotein Tah18-dependent synthesis of nitric oxide, which confers stress tolerance on S. cerevisiae cells [4][5][6]. Although other charged amino acids, such as lysine and glutamate, also effectively enhance the freeze tolerance for yeast cells [1], the mechanism underlying the freezing stress tolerance by these amino acids remained unclear. In contrast, intracellular excessive levels of basic amino acids (lysine, arginine, and histidine) in mammals were reported to induce toxicity leading to gastrointestinal diseases, such as hepatomegaly and acute pancreatitis [7][8][9]. Although pleiotropic disorders in lipid metabolism, protein synthesis, and mitochondrial functions have been observed in cells damaged by basic amino acids, their primary cellular effects are not fully understood.
–
Basic amino acids are incorporated via three similar permeases, Can1, Lyp1, and Alp1, which share 60-65% sequence identity with each other [10]. Can1 was originally identified as an arginine permease [11], though it has been also reported to transport lysine, histidine, and ornithine with lower affinities [12]. An ortholog of Can1 in Candida albicans actively transports lysine, arginine, and histidine [13]. Lyp1 efficiently mediates only lysine transport [14][15]. Overexpression of ALP1 leads to specific uptake of arginine, although it is unclear whether this gene is expressed under physiological conditions [15]. While these permeases transport amino acids with substrate preferences, the general amino-acid permease Gap1 is a transporter for all of 20 L-forms and also D-forms of the common α-amino acids, as well as other related compounds, such as citrulline, ornithine, γ-aminobutyric acid (GABA), and polyamines [16][17][18][19]. Gap1 is most closely related to Hip1 in terms of amino acid sequence, although Hip1 seems to be a rather specific permease for histidine [15][20][21]. Recent comprehensive studies revealed that single overexpression of these permeases decreases the growth rate [22][23], suggesting that S. cerevisiae can be utilized as a model to analyze the cytotoxicity caused by excess basic amino acids.
–
Among basic amino acids, histidine is especially related to copper transport [24]. Since the discovery of copper(II)-bis(L-histidinato) complex in human blood [25], extensive research has been performed to determine its physiological roles. Consequently, histidine was found to facilitate copper uptake in hepatic, placental, and brain cells [26][27][28] by removing copper from albumin, which physically inhibits incorporation of copper ions [29]. The copper(II)-bis(L-histidinato) complex has thus been applied for the treatment of Menkes disease and hypertrophic cardioencephalomyopathy, both of which are closely associated with copper deficiency [30][31]. In S. cerevisiae, copper uptake is mediated by the high-affinity transporters Ctr1 and Ctr3 and a ferric/cupric reductase Fre1, which oxidizes copper(II) into usable copper(I) ions in advance of their uptake [32][33][34]. To maintain copper homeostasis, the CTR1, CTR3, and FRE1 genes are upregulated or downregulated under copper starvation or excess copper conditions, respectively, via the action of the copper-sensing transcription factor Mac1 [35][36]. Intriguingly, the mutations in the histidine biosynthetic genes of S. cerevisiae increase sensitivity to the excess amounts of copper, which is suppressed by addition of histidine [37]. This finding supports the idea that histidine might directly interact with copper ions in yeast cells to alleviate the copper toxicity.
–
To explore novel roles of free amino acids, we analyzed here the cytotoxicity caused by exogenous addition of excess basic amino acids in S. cerevisiae.
RESULTS AND DISCUSSION
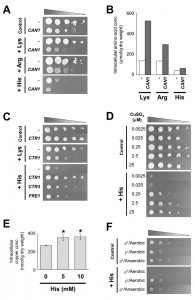
To understand the mechanism by which excess of basic amino acids mediate cytotoxicity, we examined cell growth of S. cerevisiae under culture conditions supplemented with an elevated concentration of lysine, arginine, or histidine. As shown in Figure 1A, 5 mM of histidine severely impaired the growth of yeast cells, although a higher concentration of lysine or arginine (25 mM) did not affect growth. The concentrations for three basic amino acids were selected based on the intracellular contents of these amino acids in L5487 cells (Figure 1B). When the CAN1 gene, encoding basic amino acids permease on the plasma membrane [12][15][38], was overexpressed, there was little effect on growth on SCGal medium that contained no excess of basic amino acids. In contrast, the overexpression of CAN1 markedly inhibited growth under elevated levels of basic amino acids. In particular, the growth of yeast cells that overexpress CAN1 was relatively slow on the medium in the presence of 5 mM of histidine. We confirmed that the overexpression of CAN1 increased the intracellular levels of basic amino acids in SCGal medium (approximately 1.6- to 4.1-fold increase) (Figure 1B). Thus, these results suggest that the excess amount of intracellular basic amino acids exerts toxic effects on yeast cells. Considering that Can1 preferentially transports lysine and arginine [15], overexpression of the high-affinity histidine transporter gene HIP1 [15][21] might enhance histidine uptake and effectively confer more severe toxicity to yeast cells in the presence of excess histidine. As histidine conferred more sensitivity to yeast cells than lysine and arginine despite having the lowest intracellular level, we further analyzed histidine cytotoxicity.
To identify multicopy suppressor genes that alleviate histidine toxicity, a yeast genomic library YEp51B [39] was introduced into L5487 cells overexpressing CAN1, and the transformants were screened for growth on SCGal medium containing 10 mM histidine. Sixteen independent genomic DNA fragments were isolated from the transformant colonies, and 25 full-length open reading frames were included in these fragments. After subcloning into pYES2, each gene was tested for its effect on the growth of L5487 cells in the presence of excess histidine. Consequently, the CTR1 gene, which encodes a high-affinity copper transporter that predominantly mediates copper uptake under low copper conditions [32], exhibited the most significant suppression of the histidine-caused growth defect (Figure 1C). Although the overexpression of CTR1 reduced cell growth in the presence of excess lysine by unknown mechanism(s), it is suggested that Ctr1 functions in alleviating histidine toxicity. Another high-affinity copper transporter gene, CTR3 [33], and a cupric reductase gene, FRE1, the latter of which is required for conversion of copper(II) to copper(I) ions prior to uptake [34], also suppressed the growth defect under histidine-excess conditions when overexpressed. In addition, deletion of the CTR1 or FRE1 gene slightly increased sensitivity to excess histidine (data not shown). These results indicate that the histidine cytotoxicity in S. cerevisiae is alleviated by enhancement of copper uptake. In agreement with these results, the growth defect caused by 5 mM histidine became significantly more severe when the concentration of CuSO4 in SD medium was decreased (2.5 and 25 nM), although higher concentrations of CuSO4 (2.5 and 25 µM) suppressed the histidine toxicity (Figure 1D). It has been well studied that histidine directly binds to copper(II) to form copper(II)-bis(L-histidinato) complex under physiological conditions as in human blood [24]. However, considering that the overexpression of CAN1 enhanced histidine toxicity (Figure 1A), it seems unlikely that the elevated level of histidine simply chelates copper(II) ions outside of the cells to inhibit copper uptake. Instead, excess histidine might interact with copper(I) ions to reduce the availability of copper after incorporation into yeast cells. To verify this hypothesis, we quantified the intracellular level of copper ions. Although cell growth was delayed starting after 4-hour incubation with 5 or 10 mM histidine (data not shown), intracellular copper ions were slightly increased in the histidine-treated cells (Figure 1E). Therefore, our data consistently demonstrate that an excess level of histidine enhances copper uptake but impairs copper availability in yeast cells. Regarding the CTR1-overexpressing cells (Figure 1C), sufficient copper ions might be incorporated into cells in bioavailable forms and thus contributed to relieving histidine toxicity. In a similar manner as clioquinol [40], histidine may act as both a chelator and an ionophore: histidine chelates copper(II) outside of the cell and is taken in as a complex, which may facilitate uptake of copper ions, though copper ions in this complex are not bioavailable. Additionally, it is possible that histidine-induced copper deficiency upregulates expression of copper transporters, which may increase copper uptake.
–
What functions of copper does excess histidine inhibit? Intracellular copper is distributed to distinct target proteins via specific cytosolic copper chaperons, such as Atx1, Ccs1, and Cox17. Atx1 assists in the transport of copper to the cell-surface iron uptake protein Fet3 through the function of Ccc2, which has copper-transporting ATPase activity, on the post-Golgi vesicle [41]. Ccs1 delivers copper specifically to the superoxide dismutase Sod1, which scavenges reactive oxygen species in the intermembrane space of mitochondria [42][43]. Another copper chaperon Cox17 transfers copper to the mitochondrial inner membrane proteins
–
Cox11 and Sco1, both of which are essential for assembly of cytochrome c oxidase, which is the last enzyme in the respiratory electron transport chain [44]. In this study, we tested whether histidine cytotoxicity might be mediated by defective mitochondrial functions due to reduced copper availability. As shown in Figure 1F, L5487 cells were clearly sensitive to 5 mM histidine on SD plates, although histidine cytotoxicity was completely abrogated by a spontaneous cytoplasmic petite (ρ–) mutation, which inactivates mitochondrial respiratory functions. It is also worth noting that yeast cells showed similar growth phenotypes in the absence or presence of excess histidine under anaerobic conditions (Figure 1F). Thus, histidine cytotoxicity was observed only when mitochondrial aerobic respiration should be functional. We hypothesize that the deficiency of the Cox17-bound copper ions due to excess of intracellular histidine might cause the abnormal assembly of cytochrome c oxidase complex in aerobically growing cells, leading to the observed toxic effect. This might be supported by the fact that the disturbance of cytochrome c oxidase induces apoptosis-like cell death in S. cerevisiae [45]. However, we cannot rule out the possibility that defective mitochondrial respiration reduces incorporation of histidine by some unknown mechanism(s), and hence, the excess level of histidine did not elicit the toxicity in ρ– cells under aerobic conditions or in ρ+ cells under anaerobic conditions.
–
In this study, we discovered the cytotoxicity of excess histidine in S. cerevisiae, which is tightly associated with the reduced availability of intracellular copper ions. Similarly, Pearce and Sherman [37] revealed that intracellular histidine synthesis is required for the detoxification of excess copper. Both studies commonly suggest that intracellular histidine has a novel and important role in copper homeostasis.
MATERIALS AND METHODS
Strains and Plasmids
The S. cerevisiae strain used in this study was Σ1278b wild-type strain L5487 (MATα ura3-52 leu2::hisG), which was generously provided by Gerald Fink (Whitehead Institute). A spontaneous cytoplasmic petite (ρ–) mutant of L5487 was isolated, according to Fox et al. [46]. Escherichia coli strain DH5α (F–λ–Φ80lacZ∆M15 ∆(lacZYA argF)U169 deoR recA1 endA1 hsdR17(rk–mk+) supE44 thi-1 gyrA96) was used to subclone the yeast gene and construct plasmids. Low-copy plasmids pRS415 and pRS416 [24] were used to complement auxotrophic mutations ura3 and leu2, respectively. A galactose-inducible plasmid pYES2 (Life Technologies) was used for overexpression of the CAN1, CTR1, CTR3, and FRE1 gene. A 2 µm-based yeast genomic library YEp51B [12] was used to identify multicopy suppressor genes of CAN1-overexpressing L5487 strain.
–
Culture Media
The media used for growth of S. cerevisiae were a synthetic complete medium, SC (2% glucose, 0.67% yeast nitrogen base without amino acids (Difco), supplemented with synthetic drop-out amino acid and nucleotide mixture as required), and a synthetic defined medium, SD (2% glucose, 0.67% yeast nitrogen base without amino acids (Difco)). For overexpression of CAN1, CTR1, CTR3, and FRE1, SC with galactose as a carbon source (SCGal) (2% galactose, 0.67% yeast nitrogen base without amino acids (Difco), supplemented with synthetic drop-out amino acid and nucleotide mixture as required) was used. To evaluate the effect of copper sulfate (CuSO4) addition, SD-Cu media (2% glucose, 0.67% yeast nitrogen base without amino acids and copper (ForMedium)) containing different concentrations of CuSO4 were used. All experiments were performed at 30°C, and all growth media were adjusted to pH 6.5 with HEPES buffer (pH 7.0) and sodium hydroxide. When necessary, 2% agar was added to solidify the medium. For anaerobic cultivation, the inoculated plates were incubated with O2-absorber/CO2-generator AnaeroPouch-Anaero (Mitsubishi Gas Chemical Company) and O2 indicators. The E. coli recombinant strains were grown in Luria-Bertani complete medium containing 50 µg/ml ampicillin or M9 minimal medium plus 2% Casamino acids containing 50 µg/ml ampicillin. If necessary, 2% agar was added to solidify the medium.
Measurements of Intracellular Amino Acids Levels
According to a method described previously [3], intracellular amino acids were extracted by boiling from log-phase cells cultivated in SCGal-Leu-Ura liquid medium, and were subsequently quantified with an amino acid analyzer AminoTac JLC-500/V (JEOL).
Determination of Intracellular Copper Ions
Cell lysates were prepared from log-phase cells cultivated in SC-Leu-Ura-Lys-Arg-His liquid medium, and copper ions concentrations were determined by the CUPRAC-BCS assay [25].
References
- H. Takagi, F. Iwamoto, and S. Nakamori, "Isolation of freeze-tolerant laboratory strains of Saccharomyces cerevisiae from proline-analogue-resistant mutants", Applied Microbiology and Biotechnology, vol. 47, pp. 405-411, 1997. http://dx.doi.org/10.1007/s002530050948
- Y. Morita, S. Nakamori, and H. Takagi, "Effect of proline and arginine metabolism on freezing stress of Saccharomyces cerevisiae", Journal of Bioscience and Bioengineering, vol. 94, pp. 390-394, 2002. http://dx.doi.org/10.1016/S1389-1723(02)80214-6
-
Y. Morita, S. Nakamori, and H. Takagi, "
l -Proline Accumulation and Freeze Tolerance in Saccharomyces cerevisiae Are Caused by a Mutation in the PRO1 Gene Encoding γ-Glutamyl Kinase", Applied and Environmental Microbiology, vol. 69, pp. 212-219, 2003. http://dx.doi.org/10.1128/aem.69.1.212-219.2003 - A. Nishimura, T. Kotani, Y. Sasano, and H. Takagi, "An antioxidative mechanism mediated by the yeast N-acetyltransferase Mpr1: oxidative stress-induced arginine synthesis and its physiological role", FEMS Yeast Research, vol. 10, pp. 687-698, 2010. http://dx.doi.org/10.1111/j.1567-1364.2010.00650.x
- Y. Sasano, Y. Haitani, K. Hashida, I. Ohtsu, J. Shima, and H. Takagi, "Enhancement of the proline and nitric oxide synthetic pathway improves fermentation ability under multiple baking-associated stress conditions in industrial baker's yeast", Microbial Cell Factories, vol. 11, 2012. http://dx.doi.org/10.1186/1475-2859-11-40
- A. Nishimura, N. Kawahara, and H. Takagi, "The flavoprotein Tah18-dependent NO synthesis confers high-temperature stress tolerance on yeast cells", Biochemical and Biophysical Research Communications, vol. 430, pp. 137-143, 2013. http://dx.doi.org/10.1016/j.bbrc.2012.11.023
- J.K. Solomon, and R.L. Geison, "Effect of excess dietary L-histidine on plasma cholesterol levels in weanling rats.", The Journal of nutrition, 1978. http://www.ncbi.nlm.nih.gov/pubmed/650295
- T. Mizunuma, S. Kawamura, and Y. Kishino, "Effects of injecting excess arginine on rat pancreas.", The Journal of nutrition, 1984. http://www.ncbi.nlm.nih.gov/pubmed/6199486
- G. Biczó, P. Hegyi, S. Dósa, N. Shalbuyeva, S. Berczi, R. Sinervirta, Z. Hracskó, A. Siska, Z. Kukor, K. Jármay, V. Venglovecz, I.S. Varga, B. Iványi, L. Alhonen, T. Wittmann, A. Gukovskaya, T. Takács, and Z. Rakonczay, "The Crucial Role of Early Mitochondrial Injury in L-Lysine-Induced Acute Pancreatitis", Antioxidants & Redox Signaling, vol. 15, pp. 2669-2681, 2011. http://dx.doi.org/10.1089/ars.2011.4065
- K.H. Schmidt, E. Viebranz, L. Doerfler, C. Lester, and A. Rubenstein, "Formation of Complex and Unstable Chromosomal Translocations in Yeast", PLoS ONE, vol. 5, pp. e12007, 2010. http://dx.doi.org/10.1371/journal.pone.0012007
- M. Grenson, M. Mousset, J. Wiame, and J. Bechet, "Multiplicity of the amino acid permeases in Saccharomyces cerevisiae", Biochimica et Biophysica Acta (BBA) - General Subjects, vol. 127, pp. 325-338, 1966. http://dx.doi.org/10.1016/0304-4165(66)90387-4
- T. Cooper, "Nitrogen metabolism and Saccharomyces cerevisiae. In: Strathern JN, Jones EW, and Broach JR, editors. The molecular biology of the yeast Saccharomyces: metabolism and gene expression.", Cold Spring Harbor Laboratory, New York; pp 339-461, 1982.
- H. Sychrová, and J. Souciet, "Yeast sequencing reports. CAN1, a gene encoding a permease for basic amino acids in Candida albicans", Yeast, vol. 10, pp. 1647-1651, 1994. http://dx.doi.org/10.1002/yea.320101214
- H. Sychrova, and M.R. Chevallier, "Cloning and sequencing of the Saccharomyces cerevisiae gene LYP1 coding for a lysine‐specific permease", Yeast, vol. 9, pp. 771-782, 1993. http://dx.doi.org/10.1002/yea.320090711
- B. Regenberg, L. Düring-Olsen, M.C. Kielland-Brandt, and S. Holmberg, "Substrate specificity and gene expression of the amino-acid permeases in Saccharomyces cerevisiae", Current Genetics, vol. 36, pp. 317-328, 1999. http://dx.doi.org/10.1007/s002940050506
- M. Grenson, and B. Acheroy, "Mutations affecting the activity and the regulation of the general amino-acid permease of Saccharomyces cerevisiae", Molecular and General Genetics MGG, vol. 188, pp. 261-265, 1982. http://dx.doi.org/10.1007/bf00332685
- J. JAUNIAUX, and M. GRENSON, "GAP1, the general amino acid permease gene of Saccharomyces cerevisiae", European Journal of Biochemistry, vol. 190, pp. 39-44, 1990. http://dx.doi.org/10.1111/j.1432-1033.1990.tb15542.x
- S.C. Garcia, M.B. Moretti, E. Ramos, and A. Batlle, "Carbon and nitrogen sources regulate δ-aminolevulinic acid and γ-aminobutyric acid transport in Saccharomyces cerevisiae", The International Journal of Biochemistry & Cell Biology, vol. 29, pp. 1097-1101, 1997. http://dx.doi.org/10.1016/s1357-2725(97)00047-2
- T. Uemura, K. Kashiwagi, and K. Igarashi, "Uptake of putrescine and spermidine by Gap1p on the plasma membrane in Saccharomyces cerevisiae", Biochemical and Biophysical Research Communications, vol. 328, pp. 1028-1033, 2005. http://dx.doi.org/10.1016/j.bbrc.2005.01.064
- M. Crabeel, and M. Grenson, "Regulation of Histidine Uptake by Specific Feedback Inhibition of Two Histidine Permeases in Saccharomyces cerevisiae", European Journal of Biochemistry, vol. 14, pp. 197-204, 1970. http://dx.doi.org/10.1111/j.1432-1033.1970.tb00278.x
- J. Tanaka, and G.R. Fink, "The histidine permease gene (HIP1) of Saccharomyces cerevisiae", Gene, vol. 38, pp. 205-214, 1985. http://dx.doi.org/10.1016/0378-1119(85)90219-7
- R. Sopko, D. Huang, N. Preston, G. Chua, B. Papp, K. Kafadar, M. Snyder, S.G. Oliver, M. Cyert, T.R. Hughes, C. Boone, and B. Andrews, "Mapping Pathways and Phenotypes by Systematic Gene Overexpression", Molecular Cell, vol. 21, pp. 319-330, 2006. http://dx.doi.org/10.1016/j.molcel.2005.12.011
- K. Yoshikawa, T. Tanaka, Y. Ida, C. Furusawa, T. Hirasawa, and H. Shimizu, "Comprehensive phenotypic analysis of single‐gene deletion and overexpression strains of Saccharomyces cerevisiae", Yeast, vol. 28, pp. 349-361, 2011. http://dx.doi.org/10.1002/yea.1843
- P. Deschamps, P. Kulkarnip, and B. Gautam-Basak,M:Sarkar, "The saga of copper(II)-L-histidine.", Coord Chem Rev 249(9-10): 895-909, 2005.
- B. Sarkar, and T. Kruck, "Copper-amino acid complexes in human serum.", Academic Press, New York; pp 183-196., 1966.
- H.M. Darwish, J.C. Cheney, R.C. Schmitt, and M.J. Ettinger, "Mobilization of copper(II) from plasma components and mechanisms of hepatic copper transport.", The American journal of physiology, 1984. http://www.ncbi.nlm.nih.gov/pubmed/6696070
- D.E. Hartter, and A. Barnea, "Brain tissue accumulates 67copper by two ligand-dependent saturable processes. A high affinity, low capacity and a low affinity, high capacity process.", The Journal of biological chemistry, 1988. http://www.ncbi.nlm.nih.gov/pubmed/3335527
- A. Mas, and B. Sarkar, "Uptake of 67Cu by isolated human trophoblast cells", Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, vol. 1135, pp. 123-128, 1992. http://dx.doi.org/10.1016/0167-4889(92)90127-w
- H.J. McArdle, J.R. Guthrie, M. Ackland, and D.M. Danks, "Albumin has no role in the uptake of copper by human fibroblasts", Journal of Inorganic Biochemistry, vol. 31, pp. 123-131, 1987. http://dx.doi.org/10.1016/0162-0134(87)80057-0
- B. Sarkar, "Treatment of Wilson and Menkes Diseases", Chemical Reviews, vol. 99, pp. 2535-2544, 1999. http://dx.doi.org/10.1021/cr980446m
- L.C. Papadopoulou, C.M. Sue, M.M. Davidson, K. Tanji, I. Nishino, J.E. Sadlock, S. Krishna, W. Walker, J. Selby, D.M. Glerum, R.V. Coster, G. Lyon, E. Scalais, R. Lebel, P. Kaplan, S. Shanske, D.C. De Vivo, E. Bonilla, M. Hirano, S. DiMauro, and E.A. Schon, "Fatal infantile cardioencephalomyopathy with COX deficiency and mutations in SCO2, a COX assembly gene", Nature Genetics, vol. 23, pp. 333-337, 1999. http://dx.doi.org/10.1038/15513
- A. Dancis, D.S. Yuan, D. Haile, C. Askwith, D. Eide, C. Moehle, J. Kaplan, and R.D. Klausner, "Molecular characterization of a copper transport protein in S. cerevisiae: An unexpected role for copper in iron transport", Cell, vol. 76, pp. 393-402, 1994. http://dx.doi.org/10.1016/0092-8674(94)90345-x
- S.A. Knight, S. Labbé, L.F. Kwon, D.J. Kosman, and D.J. Thiele, "A widespread transposable element masks expression of a yeast copper transport gene.", Genes & Development, vol. 10, pp. 1917-1929, 1996. http://dx.doi.org/10.1101/gad.10.15.1917
- R. Hassett, and D.J. Kosman, "Evidence for Cu(II) Reduction as a Component of Copper Uptake by Saccharomyces cerevisiae", Journal of Biological Chemistry, vol. 270, pp. 128-134, 1995. http://dx.doi.org/10.1074/jbc.270.1.128
- S. Labbé, Z. Zhu, and D.J. Thiele, "Copper-specific Transcriptional Repression of Yeast Genes Encoding Critical Components in the Copper Transport Pathway", Journal of Biological Chemistry, vol. 272, pp. 15951-15958, 1997. http://dx.doi.org/10.1074/jbc.272.25.15951
- Y. Yamaguchi-Iwai, M. Serpe, D. Haile, W. Yang, D.J. Kosman, R.D. Klausner, and A. Dancis, "Homeostatic Regulation of Copper Uptake in Yeast via Direct Binding of MAC1 Protein to Upstream Regulatory Sequences ofFRE1 and CTR1", Journal of Biological Chemistry, vol. 272, pp. 17711-17718, 1997. http://dx.doi.org/10.1074/jbc.272.28.17711
- D.A. Pearce, and F. Sherman, "Toxicity of copper, cobalt, and nickel salts is dependent on histidine metabolism in the yeast Saccharomyces cerevisiae.", Journal of bacteriology, 1999. http://www.ncbi.nlm.nih.gov/pubmed/10438744
- K. Malínská, J. Malínský, M. Opekarová, and W. Tanner, "Visualization of Protein Compartmentation within the Plasma Membrane of Living Yeast Cells", Molecular Biology of the Cell, vol. 14, pp. 4427-4436, 2003. http://dx.doi.org/10.1091/mbc.e03-04-0221
- R. Akada, J. Yamamoto, and I. Yamashita, "Screening and identification of yeast sequences that cause growth inhibition when overexpressed", Molecular and General Genetics MGG, vol. 254, pp. 267-274, 1997. http://dx.doi.org/10.1007/s004380050415
- C. Li, J. Wang, and B. Zhou, "The metal chelating and chaperoning effects of clioquinol: insights from yeast studies.", Journal of Alzheimer's disease : JAD, 2010. http://www.ncbi.nlm.nih.gov/pubmed/21504115
- S.J. Lin, R.A. Pufahl, A. Dancis, T.V. O'Halloran, and V.C. Culotta, "A role for the Saccharomyces cerevisiae ATX1 gene in copper trafficking and iron transport.", The Journal of biological chemistry, 1997. http://www.ncbi.nlm.nih.gov/pubmed/9083054
- V.C. Culotta, L.W. Klomp, J. Strain, R.L.B. Casareno, B. Krems, and J.D. Gitlin, "The Copper Chaperone for Superoxide Dismutase", Journal of Biological Chemistry, vol. 272, pp. 23469-23472, 1997. http://dx.doi.org/10.1074/jbc.272.38.23469
- L.A. Sturtz, K. Diekert, L.T. Jensen, R. Lill, and V.C. Culotta, "A fraction of yeast Cu,Zn-superoxide dismutase and its metallochaperone, CCS, localize to the intermembrane space of mitochondria. A physiological role for SOD1 in guarding against mitochondrial oxidative damage.", The Journal of biological chemistry, 2001. http://www.ncbi.nlm.nih.gov/pubmed/11500508
- Y. Horng, P.A. Cobine, A.B. Maxfield, H.S. Carr, and D.R. Winge, "Specific Copper Transfer from the Cox17 Metallochaperone to Both Sco1 and Cox11 in the Assembly of Yeast Cytochrome c Oxidase", Journal of Biological Chemistry, vol. 279, pp. 35334-35340, 2004. http://dx.doi.org/10.1074/jbc.m404747200
- X. Zuo, J.T. Djordjevic, J. Bijosono Oei, D. Desmarini, S.D. Schibeci, K.A. Jolliffe, and T.C. Sorrell, "Miltefosine Induces Apoptosis-Like Cell Death in Yeast via Cox9p in Cytochrome c Oxidase", Molecular Pharmacology, vol. 80, pp. 476-485, 2011. http://dx.doi.org/10.1124/mol.111.072322
- T.D. Fox, L.S. Folley, J.J. Mulero, T.W. McMullin, P.E. Thorsness, L.O. Hedin, and M.C. Costanzo, "Analysis and manipulation of yeast mitochondrial genes.", Methods in enzymology, 1991. http://www.ncbi.nlm.nih.gov/pubmed/1706458
ACKNOWLEDGMENTS
We are grateful to Dr. Gerald R. Fink and the NBRP of the MEXT, Japan, for providing the S. cerevisiae Σ1278b wild-type strain L5487 and a high-copy yeast genomic DNA library YEp51B, respectively.
COPYRIGHT
© 2014

Exogenous addition of histidine reduces copper availability in the yeast Saccharomyces cerevisiae by Daisuke Watanabe et al. is licensed under a Creative Commons Attribution 4.0 International License.