Research Reports:
Microbial Cell, Vol. 2, No. 7, pp. 247 - 255; doi: 10.15698/mic2015.07.213
Human Thyroid Cancer-1 (TC-1) is a vertebrate specific oncogenic protein that protects against copper and pro-apoptotic genes in yeast
1 Department of Chemistry and Chemical Engineering, Royal Military College of Canada, Kingston, Ontario, Canada.
2 Department of Anatomy and Cell Biology, McGill University, Montreal, Quebec, Canada.
3 Department of Biology, Queen’s University, Kingston, Ontario, Canada.
4 Present address: Department of Experimental Medicine, McGill University, Montreal, Quebec, Canada.
5 Present address: Department of Biomedical Sciences, Queen’s University, Kingston, Ontario, Canada.
6 Present address: Defence Research and Development Canada, Alberta, Canada.
# These authors contributed equally.
Keywords: TC-1, apoptosis, cell death, anti-apoptotic, cell survival, cancer, yeast.
Abbreviations:
hTC-1 - human Thyroid Cancer-1 protein,
PCD - programmed cell death,
ROS - reactive oxygen species.
Received originally: 10/01/2015 Received in revised form: 12/05/2015
Accepted: 08/06/2015
Published: 06/07/2015
Correspondence:
Michael T. Greenwood, Royal Military College of Canada, Department of Chemistry and Chemical Engineering, PO Box 17000 Station Forces; Kingston, Ontario, Canada, K7K 7B4 michael.greenwood@rmc.ca
Conflict of interest statement: The authors declare there is no conflict of interest.
Please cite this article as: Natalie K. Jones, Nagla T.T. Arab, Rawan Eid, Nada Gharib, Sara Sheibani, Hojatollah Vali, Chamel Khoury, Alistair Murray, Eric Boucher, Craig A. Mandato, Paul G. Young and Michael T. Greenwood (2015). Human Thyroid Cancer-1 (TC-1) is a vertebrate specific oncogenic protein that protects against copper and pro-apoptotic genes in yeast. Microbial Cell 2(7): 247-255.
Abstract
The human Thyroid Cancer-1 (hTC-1) protein, also known as C8orf4 was initially identified as a gene that was up-regulated in human thyroid cancer. Here we show that hTC-1 is a peptide that prevents the effects of over-expressing Bax in yeast. Analysis of the 106 residues of hTC-1 in available protein databases revealed direct orthologues in jawed-vertebrates, including mammals, frogs, fish and sharks. No TC-1 orthologue was detected in lower organisms, including yeast. Here we show that TC-1 is a general pro-survival peptide since it prevents the growth- and cell death-inducing effects of copper in yeast. Human TC-1 also prevented the deleterious effects that occur due to the over-expression of a number of key pro-apoptotic peptides, including YCA1, YBH3, NUC1, and AIF1. Even though the protective effects were more pronounced with the over-expression of YBH3 and YCA1, hTC-1 could still protect yeast mutants lacking YBH3 and YCA1 from the effects of copper sulfate. This suggests that the protective effects of TC-1 are not limited to specific pathways or processes. Taken together, our results indicate that hTC-1 is a pro-survival protein that retains its function when heterologously expressed in yeast. Thus yeast is a useful model to characterize the potential roles in cell death and survival of cancer related genes.
INTRODUCTION
Programmed cell death (PCD) is a biological process that is activated in response to a variety of stresses [1][2]. Stresses can serve to activate both pro- and anti-apoptotic processes in order to destroy the cell or to prevent the cell from undergoing a pre-mature death [3]. The most commonly studied form of genetically encoded cell death is controlled by the Bcl-2 family of proteins [4][5]. Bax is a pro-death Bcl-2 family member that gets activated by a variety of stresses in order to initiate cell death processes. In apoptotic cell death, Bax leads to mitochondrial damage, increases in reactive oxygen species (ROS), release of apoptogenic mitochondrial factors including cytochrome c, AIF and EndoG that lead to controlled cell breakdown in part by the activation of caspases [6]. The effects of activated Bax can be offset by the pro-survival Bcl-2 protein. In addition to inhibiting Bax, overexpressed Bcl-2 can prevent multiple forms of cell death including cell death that occurs in the absence of activated Bax [7]. The ability to prevent multiple forms of cell death is a commonly observed for many pro-survival proteins [3].
–
Programmed cell death can occur via at least three different processes including apoptosis (type I), necrosis or necroptosis (type II) and autophagy (type III) [2][6]. The complexity of PCD is further enhanced by the fact that there are multiple sub-forms of all PCD sub-types as well as by the existence of extensive cross-talk between the different forms [1][2][3]. Many human diseases, such as ischemic heart and neurological diseases, are associated with abnormal signaling within the cell, which gives rise to altered levels of cell death. This has led to the development of therapeutic strategies aimed at increasing or decreasing the function of apoptotic regulators [4]. Blocking specific apoptotic effectors like caspases have been shown to have limited success in blocking cell death in in vivo models [3][8]. This is likely due to the activation of alternative cell death pathways or to the inability to interfere with cell death once the process has been initiated. Initially, apoptosis was thought to be the predominant form of cell death in many diseases, but it is now clear that other forms of cell death including necroptosis play important roles in the loss of cells [9][10][11]. In contrast, other diseases such as cancer are associated with decreases in cell death responses [12][13][14]. Increases in the protective effects of autophagy as well as increases in the expression levels of pro-survival proteins, such as Bcl-2, can promote changes in the apoptotic and PCD responses [12][15]. Thus, instead of developing chemicals targeting specific apoptotic regulators, the development of strategies based on the knockdown or the overexpression of apoptotic regulators is being developed as possible therapies for cancer and other diseases [16][17]. The development of model transgenic animals overexpressing pro-survival peptides supports such strategies [18].
–
Following the initial identification of a PCD pathway in the worm C. elegans, it became clear that PCD was conserved in multicellular organisms [6][19]. The identification of conserved pathways in single celled organisms, including numerous species of yeast, has led to novel insights into the diversity of PCD pathways [20]. More elaborate insights into the basic PCD processes have been uncovered in extensive studies that have now been carried out using the genetically tractable yeast Saccharomyces cerevisiae [18][21][22][23]. In effect, yeast has proven useful for the study of all sub-forms of PCD [21][24][25][26][27]. Yeast undergo PCD in response to a variety of different stresses and they display the typical hallmarks of cell death that can be easily monitored [25]. Yeast also have apparent functional orthologues to a number of mammalian cell death regulators including caspase (MCA1), AIF, EndoG (Nuc1) and YBH3, a single BH3 domain Bcl-2 protein [28][29][30][31][32]. So it is not surprising that yeast has proven to be an effective model for PCD processes including many neurological diseases such as Huntington’s and Parkinson’s disease [23]. Thus the expression of human α-synuclein leads to cell death in yeast in a manner that is analogous to what is seen in Parkinson’s [33]. More recent studies have suggested that increased calcium, mitophagy and specific genes including EndoG may be involved in mediating the toxicity of α-synuclein [30][34][35]. Further humanized yeast is proving to be an excellent model to screen for drugs that can prevent cell death in response to PCD inducing conditions, including the heterologous expression of specific human genes [36][37][38]. Similarly, yeast has proven effective in examining the effects of specific cancer drugs or to screen for drugs that may be therapeutically useful as chemotherapeutics [39].
–
We and others have used yeast conditionally expressing mammalian pro-apoptotic Bax as a model system to study PCD and as a platform to screen for and identify novel negative regulators [40][41][42][43][44][45][46]. Many of the pro-survival genes identified in these heterologous screens have been shown to be pro-survival in their native mammalian hosts [40][47]. Here we have identified a cDNA encoding human Thyroid Cancer-1 (hTC-1) [48] as a peptide capable of preventing the inhibitory effects of Bax in yeast. We further show that this peptide retains its pro-survival functions in yeast in response to other stresses including copper and activated pro-apoptotic peptides. These results are consistent with the observation that TC-1 is up-regulated by and serves to inhibit stress and apoptosis [49][50][51]. Thus, yeast is a useful model to characterize potential cell survival functions of genes that are up-regulated in cancer.
RESULTS AND DISCUSSION
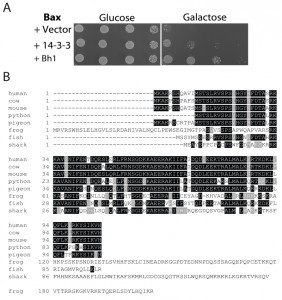
In a previous screen of a human cardiac cDNA library, Bh-1 was identified as a Bax suppressor [52]. The Bh-1-expressing plasmid was isolated from the original yeast transformant and retransformed, along with the Bax expressing plasmid into the wild type strain. Cultures of cells harbouring Bax and the empty vector, Bax and 14-3-3β/α or Bax with Bh-1, were serially diluted and spotted onto nutrient agar media with glucose or galactose (Figure 1A). The human 14-3-3β/α cDNA was used as a control as it had been previously identified as an anti-apoptotic peptide [53]. In Figure 1A, all three transformants show the same growth on glucose media. In contrast, cells harbouring Bax and the vector alone showed reduced growth when spotted onto the galactose-inducing media, as the pro-apoptotic Bax-expressing plasmid was activated. The co-expression of the Bh1-peptide along with the Bax (Bax+Bh-1) showed enhanced cell growth on the galactose media, indicating that Bh-1 is capable of suppressing the effects of Bax. The 0.8 kb Bh1 sequence contained an entire open reading frame from nucleotide 34 to 351 that was predicted to encode a 106 residue protein. The peptide’s sequence was compared to the proteins in the GenBank human database at NCBI. The sequence is identical to the 106 residue protein that is encoded for by a 1,841 bp cDNA (with a coding sequence between nt 66-386), corresponding to the human Thyroid Cancer-1 (TC-1) gene, also known as chromosome 8 open reading frame 4 (C8orf4) (Accession Number: NM_020130). Thus, the Bh-1 sequence corresponds to human TC-1.
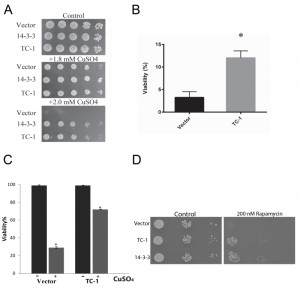
 | FIGURE 1: Identification of human TC-1 as a Bax suppressor that protects against programmed cell death-inducing levels of copper.
(A) Plasmid DNA with the Bh1 sequence was obtained from the clone identified from the original cDNA library screen for Bax suppressors [52]. It was then reintroduced into naïve BY4742 yeast cells with and without Bax. A human 14-3-3β/α cDNA was used as a positive control [53]. Cultures of the transformants were grown, serially diluted, and spotted onto YNB media with glucose or with galactose. The plates were allowed to grow at 30°C and photographs of the plates are shown.
(B) The amino acid sequence of human TC-1 was used to search the available protein databases at NCBI. Boxshade of the TCoffee alignment of representative sequences from different species showing strong similarity with hTC-1 are shown. The black shading indicates identical residues while grey indicates conserved residues. The scientific names and database accession numbers of the species used are as follows: human: Homo sapiens (Accession Number NM_020130); cow: Bos taurus (Accession Number NP_001030567); mouse: Mus musculus (Accession Number NP_081207); python: Python bivittatus (Accession Number XP_007430414.1); pigeon: Columba livia (Accession Number XP_005510897.1); frog: Xenopus tropicalis (Accession Number XP_002936483.2); fish: Poecilia formosa (Accession Number XP_007573497.1); and shark: Callorhinchus milii (Accession Number XP_007883063.1). |
In order to identify possible orthologues of the human TC-1 gene, the sequence was compared to the protein sequences available in the GenBank database. TC-1 peptides were found in multiple mammalian species as well as lower vertebrates including birds, reptiles, amphibians, sharks and fish. A representative sequence from each group was selected and these were then aligned with TCoffee and reformatted with BoxShade (http://www.ch.embnet.org/software/BOX_form.html) [54] (Figure 1B). The similarity between human TC-1 is nearly identical over the entire protein to other mammalian species [55]. The similarity subsequently decreases further away from human, with the most divergent species from human, but still containing the TC-1 domain, being the elephant shark (37% identity over 80 residues). Earlier chordates, such as lampreys and hagfishes, were also searched for sequences having similarity to TC-1. However, the search returned sequences that had very little similarity. The last similar species, the shark, belongs to the jawed-vertebrates (gnathostomes- a sub-phylum of the vertebrata), which are known for having jaws, several types of mineralized bone, an adaptive immune system and limb development. This indicates that the introduction of the TC-1 gene appears to be between the divergence of the agnatha and the gnathostomata of the vertebrata phylum (550-450 MYA) [56].
–
In order to determine the most similar yeast proteins to TC-1, the yeast and fungal databases from www.yeastgenome.org were examined. It revealed three proteins encoded by YGR238C, YGR198W and YIR001C, which are suspected to negatively regulate mitotic exit, affect cargo transportation in endocytosis and bind to RNA in the cytoplasm, respectively. From these, the strongest case for a TC-1-like sequence is with YIR001Cp, an RNA-binding protein that modulates cytoplasmic mRNA. This protein is of comparable size, as it is 245 amino acids, while the other two are 882 and 817 amino acids, respectively. The sequence of YIR001Cp has limited similarity from residues 1 to 90 with the human protein (11 out of 90 identical and 16 out of 90 with conserved amino acids). It therefore seems unlikely that the yeast protein, YIR001Cp is a TC-1 protein. It remains possible that a functional TC-1 orthologue exists in yeast, but this sequence diverged so much from its ancestral TC-1 that it does not have very direct sequence conservation with TC-1. Discerning subtle similarities between evolutionary distant ancestors has been successful in identifying a number of yeast orthologues to key mammalian apoptotic regulators, including YBH3 and YCA1 [28][29].
–
Given that alternative splicing is a common occurrence in genes encoding apoptotic regulators [57][58], we also examined the human TC-1 gene structure, with regards to its intron and exon organization. The TC-1 mRNA sequence was compared to the entire human genome, using the available database at NCBI. The analysis returned one continuous piece of DNA containing the entire TC-1 cDNA located on chromosome eight (Accession Number: AC_000140.1, between position 38544322 to 38546161 bp). The gene and cDNA sequences were continuous with each other, indicating that the entire cDNA was encoded for by a single exon. Therefore, TC-1 does not appear to have any splice-variants.
–
In order to determine whether TC-1 could protect against other stressors besides Bax, cell-death-inducing levels of copper sulfate were used [59]. Using the spot assay with increasing concentrations of copper sulfate, the cells harbouring only the vector show reduced growth, while cells harbouring TC-1 and 14-3-3 expressing plasmids remain largely unaffected (Figure 2A). In order to assess the ability of cells to survive exposure to stress and remain viable, cells were exposed to copper sulfate in liquid media for 20 h and their viability was determined using a clonogenicity assay. The viability of the empty vector control cells was reduced to 3.3 ± 2.2%, whereas the viability of cells expressing TC-1 was significantly higher at 12 ± 2.7%, an approximate 4-fold increase in viability in comparison to the vector (Figure 2B). We also carried the same experiment and directly evaluated viability after copper treatment by microscopical examination of cells stained with the vital dye trypan blue. In this assay, the viability of control cells decreased to 29.3 ± 2.1% with copper while the viability of TC-1 expressing cells was not as affected by copper as the viability was found to be 71.8 ± 1.9% (Figure 2C).
Although both methodologies yield similar results, there are notable differences in the apparent viabilities using clonogenicity and vital dye assays. This reflects differences in the timing and of the end points monitored as discussed in [57][58]. Finally we evaluated the general nature of the pro-survival function of TC-1 by examining its ability to prevent the effects of rapamycin. Rapamycin is a well known inducer of autophagy that also induces PCD, that resembles apoptosis [53]. Using the spot assay we show that control cells show reduced ability to grow on nutrient agar media that contains rapamycin (Figure 2D). As previously described, human 14-3-3 can confer partial resistance to rapamycin [53]. Cells expressing TC-1 also have increased resistance to rapamycin indicating that TC-1 can protect cells from a variety of different stresses. Taken together these results indicate that TC-1 is capable of preventing stress-mediated cell death. Further the pro-survival properties of hTC-1 in yeast are consistent with the known stress-reducing and anti-apoptotic properties of this protein in mammalian cells [49][50][51]. Thus yeast is a unique platform that may be useful in delineating the mechanisms by which TC-1 protects cells from stress. This is of potential clinical value since hTC-1 is reported to be transformative and may be a key tumor promoter in certain cancers [51][60][61][62]. Thus yeast expressing hTC-1 may also serve as a platform to screen for novel inhibitors that may have therapeutic value [39].
–
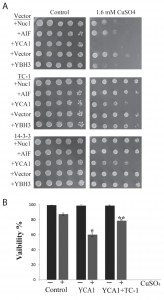
There is a number of common processes that get activated by stress in yeast and mammalian cells that occur in response to the activation of specific pro-apoptotic proteins [21][25][63]. To determine possible specific functions of TC-1, we generated yeast transformants that overexpress key pro-apoptotic peptides including NUC1, AIF, YCA1 and YBH3 with and without TC-1. NUC1 encodes Nuclease 1 (EndoG), a protein known to translocate from the mitochondria to the nucleus in order to degrade DNA under stressful conditions [64], while AIFp is Apoptosis-Inducing Factor, a protein known to be released from the mitochondria under stressful conditions and mediates cell death by degrading DNA [31]. YCA1p (also called MCA1p) is Yeast Caspase-1, a metacaspase that regulates apoptosis upon induction of a stress such as hydrogen peroxide and acetic acid [29]. YBH3p is Yeast Bax Homology 3 protein that is a Bcl-2 family protein and induces PCD [28]. Sub-lethal level of a mild stress is often required to activate over-expressed native pro-apoptotic proteins for them to induce cell death. Here we used copper sulfate at a concentration that is known to be sub-lethal [59]. Cultures of yeast doubly transformed with different combinations of expression vectors, including empty vectors, pro-apoptotic genes and TC-1 or 14-3-3 were serially diluted and spotted on nutrient agar media containing galactose alone or with sub-lethal copper sulfate (Figure 3A). The inhibitory effects observed with all four pro-apoptotic genes, was compensated for by the over-expression of TC-1 or 14-3-3 (Figure 3A). Thus, hTC-1 was able to protect against the effects of multiple pro-apoptotic genes. We confirmed the results of the spot assays using vital dye based assays. Cultures of yeast cells having both empty vectors, empty vector and YCA1 overexpressing plasmid as well as yeast having YCA1 and TC-1 expressing plasmids were challenged with low level of copper in order to activate the overexpressed Yca1p and viability was determined 18 hours later. Yeast expressing YCA1 had a lower survival rate (62.0 ± 6.2%), compared to control cells with both empty vectors (87.4 ± 2.8%) (Figure 3B). This reduced viability, induced by activated Yca1p was mitigated by the co-expression of TC-1 as cells expressing both TC-1 and YCA1 had increased survival rates (78.8 ±0.5%), compared to yeast expressing YCA1 alone (62.0 ±6.2%) (Figure 3B).
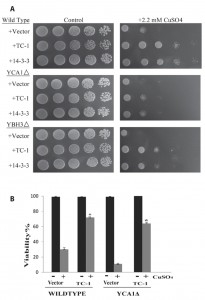
In our hands the overexpressed YCA1 and YBH3 were the most lethal and their effects were reversed by TC-1 (Figure 3A). In order to examine the possibility that TC-1 may be inhibiting a specific YCA1p and/or YBH3p pathway, we investigated if the knockout of YBH3 or YCA1 would affect the ability of TC-1 to protect the cell. Cultures of wild type as well as YCA1Δ and YBH3Δ mutants harbouring the vector, TC-1, or 14-3-3-expressing plasmids, were spotted onto nutrient agar plates with and without copper stress (Figure 4A). TC-1 was able to overcome the inhibitory effects of copper in all three strains.
–
We have previously reported that cells lacking YCA1 are more sensitive to copper than wild type cells [65]. This increased sensitivity to copper, seen in Figure 4A, serves to decrease the apparent protective effect of TC-1. We thus used vital dye to further examine the effect of TC-1 on copper mediated cell death in YCA1Δ cells. The viability of YCA1Δ cells treated with copper is decreased to 11.1 ± 1.7% while the viability of these mutant cells is increased to 63.7 ± 2.9% by the expression of TC-1 (Figure 4B). Taken together these results indicate that TC-1 is not specific to YCA1p or YBH3p pathways and may be indicative that TC-1 is acting downstream of many different pro-apoptotic proteins.
Other studies have also seen the protective effects of TC-1. It was demonstrated that TC-1 binds to the β-cantenin binding nuclear protein Chibby. Given that Chibby is a negative regulator of β-cantenin, TC-1 thus enhances Wnt pathway and increases cell proliferation [66]. However, yeast is not known to have a Wnt-signaling pathway, indicating that the human TC-1 gene is capable of activating an additional pathway that is present in yeast [67]. Another possibility is that TC-1 may act as a regulator of the heat shock response [49]. This may indicate why TC-1 is capable of protecting against a myriad of pro-apoptotic stresses and proteins, and why some of the effects observed of TC-1 are similar to the effects of 14-3-3, a chaperone type protein [53].
MATERIALS AND METHODS
Yeast Strains, Plasmids and Growth
The Saccharomyces cerevisiae BY4742 (MATα his3∆1 leu2∆0 lys15∆0 ura3∆0) strain was used for the wild type strain. The YCA1∆ and YBH3∆ deletion strains were isogenic to BY4742 [59]. The plasmids expressing the cDNAs for human 14-3-3β/α and TC-1 (Bh1) as well as yeast YBH3, YCA1, AIF1 and YBH3 expressed under the control of the GAL1-galactose-inducible promoter, were previously isolated and described [28][29][30][31][52][53]. The plasmids were introduced into yeast using the lithium acetate method and were selected for, and maintained by, the omission of the appropriate nutrient on YNBD (1% yeast nitrogen base (YNB), 2% glucose and the required amino acids). In order to express the plasmid under the influence of the GAL1 promoter, yeast were grown on YNBGal/Raf (1% YNB, 1% raffinose and 2% galactose).
–
Spot and Viability Assays
Yeast transformants that were maintained on YNBD media with supplements, were used to inoculate liquid YNBD and were incubated at 30°C for 18–24 h with shaking. The freshly saturated cultures were then used to inoculate liquid YNBGal/Raf (1 in 20) and incubated at 30°C for 4–6 h with shaking. The cultures were then serially diluted (1:5) with sterile distilled water and 10 μL of each dilution was spotted onto agar plates, and subsequently incubated at 30°C for three days. Spot assays were repeated for a minimum of three times. Any substance, such as copper sulfate, was sterilized before its addition to the agar plates. For viability assays, freshly saturated cultures were then used to inoculate liquid YNBGal/Raf (1 in 100) and incubated at 30°C with shaking with 1.4 or 1.6 mM copper sulfate (CuSO4) was added to the cultures and then incubated at 30°C for 18-20 h with shaking. The cultures were then serially diluted, spread onto nutrient agar plates, incubated at 30°C for 48 h and the number of colonies determined [58]. Viability assays were repeated for a minimum of three times and a minimum one hundred colonies were counted for each assay. Alternatively the cells were stained with the vital dye for 10 minutes then viability was determined by microscopical examination of at least 300 cells in three different experiments [59]. In order to activate the proteins that are overexpressed from the different pro-apoptotic genes, the cells were treated with sub-lethal levels of copper (1.6 mM for agar plates and 1.0 mM for liquid cultures).
–
Statistical Analysis
The two-tail one-way ANOVA was performed using GraphPad by Prism in order to assess for statistical significance.
References
- L. Galluzzi, I. Vitale, J.M. Abrams, E.S. Alnemri, E.H. Baehrecke, M.V. Blagosklonny, T.M. Dawson, V.L. Dawson, W.S. El-Deiry, S. Fulda, E. Gottlieb, D.R. Green, M.O. Hengartner, O. Kepp, R.A. Knight, S. Kumar, S.A. Lipton, X. Lu, F. Madeo, W. Malorni, P. Mehlen, G. Nuñez, M.E. Peter, M. Piacentini, D.C. Rubinsztein, Y. Shi, H. Simon, P. Vandenabeele, E. White, J. Yuan, B. Zhivotovsky, G. Melino, and G. Kroemer, "Molecular definitions of cell death subroutines: recommendations of the Nomenclature Committee on Cell Death 2012", Cell Death & Differentiation, vol. 19, pp. 107-120, 2011. http://dx.doi.org/10.1038/cdd.2011.96
- V. Nikoletopoulou, M. Markaki, K. Palikaras, and N. Tavernarakis, "Crosstalk between apoptosis, necrosis and autophagy", Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, vol. 1833, pp. 3448-3459, 2013. http://dx.doi.org/10.1016/j.bbamcr.2013.06.001
- L. Portt, G. Norman, C. Clapp, M. Greenwood, and M.T. Greenwood, "Anti-apoptosis and cell survival: A review", Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, vol. 1813, pp. 238-259, 2011. http://dx.doi.org/10.1016/j.bbamcr.2010.10.010
- M. Kvansakul, and M.G. Hinds, "The Bcl-2 family: structures, interactions and targets for drug discovery", Apoptosis, vol. 20, pp. 136-150, 2014. http://dx.doi.org/10.1007/s10495-014-1051-7
- J.K. Brunelle, and A. Letai, "Control of mitochondrial apoptosis by the Bcl-2 family", Journal of Cell Science, vol. 122, pp. 437-441, 2009. http://dx.doi.org/10.1242/jcs.031682
- N. Maghsoudi, Z. Zakeri, and R.A. Lockshin, "Programmed cell death and apoptosis--where it came from and where it is going: from Elie Metchnikoff to the control of caspases.", Experimental oncology, 2012. http://www.ncbi.nlm.nih.gov/pubmed/23069998
- M. Germain, and R. Slack, "Dining in with BCL-2: new guests at the autophagy table", Clinical Science, vol. 118, pp. 173-181, 2009. http://dx.doi.org/10.1042/CS20090310
- L. Galluzzi, J.M. Bravo-San Pedro, I. Vitale, S.A. Aaronson, J.M. Abrams, D. Adam, E.S. Alnemri, L. Altucci, D. Andrews, M. Annicchiarico-Petruzzelli, E.H. Baehrecke, N.G. Bazan, M.J. Bertrand, K. Bianchi, M.V. Blagosklonny, K. Blomgren, C. Borner, D.E. Bredesen, C. Brenner, M. Campanella, E. Candi, F. Cecconi, F.K. Chan, N.S. Chandel, E.H. Cheng, J.E. Chipuk, J.A. Cidlowski, A. Ciechanover, T.M. Dawson, V.L. Dawson, V. De Laurenzi, R. De Maria, K. Debatin, N. Di Daniele, V.M. Dixit, B.D. Dynlacht, W.S. El-Deiry, G.M. Fimia, R.A. Flavell, S. Fulda, C. Garrido, M. Gougeon, D.R. Green, H. Gronemeyer, G. Hajnoczky, J.M. Hardwick, M.O. Hengartner, H. Ichijo, B. Joseph, P.J. Jost, T. Kaufmann, O. Kepp, D.J. Klionsky, R.A. Knight, S. Kumar, J.J. Lemasters, B. Levine, A. Linkermann, S.A. Lipton, R.A. Lockshin, C. López-Otín, E. Lugli, F. Madeo, W. Malorni, J. Marine, S.J. Martin, J. Martinou, J.P. Medema, P. Meier, S. Melino, N. Mizushima, U. Moll, C. Muñoz-Pinedo, G. Nuñez, A. Oberst, T. Panaretakis, J.M. Penninger, M.E. Peter, M. Piacentini, P. Pinton, J.H. Prehn, H. Puthalakath, G.A. Rabinovich, K.S. Ravichandran, R. Rizzuto, C.M. Rodrigues, D.C. Rubinsztein, T. Rudel, Y. Shi, H. Simon, B.R. Stockwell, G. Szabadkai, S.W. Tait, H.L. Tang, N. Tavernarakis, Y. Tsujimoto, T. Vanden Berghe, P. Vandenabeele, A. Villunger, E.F. Wagner, H. Walczak, E. White, W.G. Wood, J. Yuan, Z. Zakeri, B. Zhivotovsky, G. Melino, and G. Kroemer, "Essential versus accessory aspects of cell death: recommendations of the NCCD 2015", Cell Death & Differentiation, vol. 22, pp. 58-73, 2014. http://dx.doi.org/10.1038/cdd.2014.137
- N. Vanlangenakker, T. Vanden Berghe, and P. Vandenabeele, "Many stimuli pull the necrotic trigger, an overview", Cell Death & Differentiation, vol. 19, pp. 75-86, 2011. http://dx.doi.org/10.1038/cdd.2011.164
- G. Kung, K. Konstantinidis, and R.N. Kitsis, "Programmed Necrosis, Not Apoptosis, in the Heart", Circulation Research, vol. 108, pp. 1017-1036, 2011. http://dx.doi.org/10.1161/CIRCRESAHA.110.225730
- L. Galluzzi, T. Vanden Berghe, N. Vanlangenakker, S. Buettner, T. Eisenberg, P. Vandenabeele, F. Madeo, and G. Kroemer, "Programmed Necrosis", International Review of Cell and Molecular Biology, pp. 1-35, 2011. http://dx.doi.org/10.1016/B978-0-12-386039-2.00001-8
- S. Fulda, "Evasion of Apoptosis as a Cellular Stress Response in Cancer", International Journal of Cell Biology, vol. 2010, pp. 1-6, 2010. http://dx.doi.org/10.1155/2010/370835
- A.K. Biala, and L.A. Kirshenbaum, "The interplay between cell death signaling pathways in the heart", Trends in Cardiovascular Medicine, vol. 24, pp. 325-331, 2014. http://dx.doi.org/10.1016/j.tcm.2014.08.002
- B. Favaloro, N. Allocati, V. Graziano, C. Di Ilio, and V. De Laurenzi, "Role of apoptosis in disease.", Aging, 2012. http://www.ncbi.nlm.nih.gov/pubmed/22683550
- A. Glory, A. Bettaieb, and D.A. Averill-Bates, "Mild thermotolerance induced at 40 °C protects cells against hyperthermia-induced pro-apoptotic changes in Bcl-2 family proteins", International Journal of Hyperthermia, vol. 30, pp. 502-512, 2014. http://dx.doi.org/10.3109/02656736.2014.968641
- L. Jia, S. Chen, and A. Yang, "Cancer gene therapy targeting cellular apoptosis machinery", Cancer Treatment Reviews, vol. 38, pp. 868-876, 2012. http://dx.doi.org/10.1016/j.ctrv.2012.06.008
- A. Strasser, S. Cory, and J.M. Adams, "Deciphering the rules of programmed cell death to improve therapy of cancer and other diseases", The EMBO Journal, vol. 30, pp. 3667-3683, 2011. http://dx.doi.org/10.1038/emboj.2011.307
- C. Clapp, L. Portt, C. Khoury, S. Sheibani, R. Eid, M. Greenwood, H. Vali, C.A. Mandato, and M.T. Greenwood, "Untangling the Roles of Anti-Apoptosis in Regulating Programmed Cell Death using Humanized Yeast Cells", Frontiers in Oncology, vol. 2, 2012. http://dx.doi.org/10.3389/fonc.2012.00059
- H.R. Horvitz, "NOBEL LECTURE: Worms, Life and Death", Bioscience Reports, vol. 23, pp. 239-303, 2003. http://dx.doi.org/10.1023/b:bire.0000019187.19019.e6
- A. Sharon, A. Finkelstein, N. Shlezinger, and I. Hatam, "Fungal apoptosis: function, genes and gene function", FEMS Microbiology Reviews, vol. 33, pp. 833-854, 2009. http://dx.doi.org/10.1111/j.1574-6976.2009.00180.x
- D. Carmona-Gutierrez, T. Eisenberg, S. Büttner, C. Meisinger, G. Kroemer, and F. Madeo, "Apoptosis in yeast: triggers, pathways, subroutines", Cell Death & Differentiation, vol. 17, pp. 763-773, 2010. http://dx.doi.org/10.1038/cdd.2009.219
- A.J. Munoz, K. Wanichthanarak, E. Meza, and D. Petranovic, "Systems biology of yeast cell death", FEMS Yeast Research, vol. 12, pp. 249-265, 2012. http://dx.doi.org/10.1111/j.1567-1364.2011.00781.x
- V. Khurana, and S. Lindquist, "Modelling neurodegeneration in Saccharomyces cerevisiae: why cook with baker's yeast?", Nature Reviews Neuroscience, vol. 11, pp. 436-449, 2010. http://dx.doi.org/10.1038/nrn2809
- M. Côrte-Real, and F. Madeo, "Yeast Programed Cell Death and Aging", Frontiers in Oncology, vol. 3, 2013. http://dx.doi.org/10.3389/fonc.2013.00283
- D. Wloch-Salamon, and A. Bem, "Types of cell death and methods of their detection in yeast Saccharomyces cerevisiae", Journal of Applied Microbiology, vol. 114, pp. 287-298, 2012. http://dx.doi.org/10.1111/jam.12024
- T. Eisenberg, D. Carmona-Gutierrez, S. Büttner, N. Tavernarakis, and F. Madeo, "Necrosis in yeast", Apoptosis, vol. 15, pp. 257-268, 2010. http://dx.doi.org/10.1007/s10495-009-0453-4
- B. Sampaio-Marques, C. Felgueiras, A. Silva, F. Rodrigues, and P. Ludovico, "Yeast chronological lifespan and proteotoxic stress: is autophagy good or bad?", Biochemical Society Transactions, vol. 39, pp. 1466-1470, 2011. http://dx.doi.org/10.1042/BST0391466
- S. Büttner, D. Ruli, F. Vögtle, L. Galluzzi, B. Moitzi, T. Eisenberg, O. Kepp, L. Habernig, D. Carmona-Gutierrez, P. Rockenfeller, P. Laun, M. Breitenbach, C. Khoury, K. Fröhlich, G. Rechberger, C. Meisinger, G. Kroemer, and F. Madeo, "A yeast BH3-only protein mediates the mitochondrial pathway of apoptosis", The EMBO Journal, vol. 30, pp. 2779-2792, 2011. http://dx.doi.org/10.1038/emboj.2011.197
- F. Madeo, E. Herker, C. Maldener, S. Wissing, S. Lächelt, M. Herlan, M. Fehr, K. Lauber, S.J. Sigrist, S. Wesselborg, and K. Fröhlich, "A Caspase-Related Protease Regulates Apoptosis in Yeast", Molecular Cell, vol. 9, pp. 911-917, 2002. http://dx.doi.org/10.1016/s1097-2765(02)00501-4
- S. Büttner, L. Habernig, F. Broeskamp, D. Ruli, F.N. Vögtle, M. Vlachos, F. Macchi, V. Küttner, D. Carmona-Gutierrez, T. Eisenberg, J. Ring, M. Markaki, A.A. Taskin, S. Benke, C. Ruckenstuhl, R. Braun, C. Van den Haute, T. Bammens, A. van der Perren, K. Fröhlich, J. Winderickx, G. Kroemer, V. Baekelandt, N. Tavernarakis, G.G. Kovacs, J. Dengjel, C. Meisinger, S.J. Sigrist, and F. Madeo, "Endonuclease G mediates α-synuclein cytotoxicity during Parkinson's disease", The EMBO Journal, vol. 32, pp. 3041-3054, 2013. http://dx.doi.org/10.1038/emboj.2013.228
- S. Wissing, P. Ludovico, E. Herker, S. Büttner, S.M. Engelhardt, T. Decker, A. Link, A. Proksch, F. Rodrigues, M. Corte-Real, K. Fröhlich, J. Manns, C. Candé, S.J. Sigrist, G. Kroemer, and F. Madeo, "An AIF orthologue regulates apoptosis in yeast", The Journal of Cell Biology, vol. 166, pp. 969-974, 2004. http://dx.doi.org/10.1083/jcb.200404138
- J. Cebulski, J. Malouin, N. Pinches, V. Cascio, and N. Austriaco, "Yeast Bax Inhibitor, Bxi1p, Is an ER-Localized Protein That Links the Unfolded Protein Response and Programmed Cell Death in Saccharomyces cerevisiae", PLoS ONE, vol. 6, pp. e20882, 2011. http://dx.doi.org/10.1371/journal.pone.0020882
- V. Franssens, E. Boelen, J. Anandhakumar, T. Vanhelmont, S. Büttner, and J. Winderickx, "Yeast unfolds the road map toward α-synuclein-induced cell death", Cell Death & Differentiation, vol. 17, pp. 746-753, 2009. http://dx.doi.org/10.1038/cdd.2009.203
- B. Sampaio-Marques, C. Felgueiras, A. Silva, M. Rodrigues, S. Tenreiro, V. Franssens, A.S. Reichert, T.F. Outeiro, J. Winderickx, and P. Ludovico, "SNCA (α-synuclein)-induced toxicity in yeast cells is dependent on Sir2-mediated mitophagy", Autophagy, vol. 8, pp. 1494-1509, 2012. http://dx.doi.org/10.4161/auto.21275
- G. Caraveo, P.K. Auluck, L. Whitesell, C.Y. Chung, V. Baru, E.V. Mosharov, X. Yan, M. Ben-Johny, M. Soste, P. Picotti, H. Kim, K.A. Caldwell, G.A. Caldwell, D. Sulzer, D.T. Yue, and S. Lindquist, "Calcineurin determines toxic versus beneficial responses to α-synuclein", Proceedings of the National Academy of Sciences, vol. 111, 2014. http://dx.doi.org/10.1073/pnas.1413201111
- D.F. Tardiff, V. Khurana, C.Y. Chung, and S. Lindquist, "From yeast to patient neurons and back again: Powerful new discovery platforms", Movement Disorders, vol. 29, pp. 1231-1240, 2014. http://dx.doi.org/10.1002/mds.25989
- S. Ju, D.F. Tardiff, H. Han, K. Divya, Q. Zhong, L.E. Maquat, D.A. Bosco, L.J. Hayward, R.H. Brown, S. Lindquist, D. Ringe, and G.A. Petsko, "A Yeast Model of FUS/TLS-Dependent Cytotoxicity", PLoS Biology, vol. 9, pp. e1001052, 2011. http://dx.doi.org/10.1371/journal.pbio.1001052
- R. D. Silva, S. Manon, J. Goncalves, L. Saraiva, and M. Corte-Real, "The Importance of Humanized Yeast to Better Understand the Role of Bcl-2 Family in Apoptosis: Finding of Novel Therapeutic Opportunities", Current Pharmaceutical Design, vol. 17, pp. 246-255, 2011. http://dx.doi.org/10.2174/138161211795049651
- R. Matuo, F.G. Sousa, D.G. Soares, D. Bonatto, J. Saffi, A.E. Escargueil, A.K. Larsen, and J.A.P. Henriques, "Saccharomyces cerevisiae as a model system to study the response to anticancer agents", Cancer Chemotherapy and Pharmacology, vol. 70, pp. 491-502, 2012. http://dx.doi.org/10.1007/s00280-012-1937-4
- I.S. Woo, H. Jang, S.Y. Eun, H.J. Kim, S.A. Ham, H.J. Kim, J.H. Lee, K.C. Chang, J. Kim, C.W. Han, and H.G. Seo, "Ran suppresses paclitaxel-induced apoptosis in human glioblastoma cells", Apoptosis, vol. 13, pp. 1223-1231, 2008. http://dx.doi.org/10.1007/s10495-008-0247-0
- I. Kiššová, L. Plamondon, L. Brisson, M. Priault, V. Renouf, J. Schaeffer, N. Camougrand, and S. Manon, "Evaluation of the Roles of Apoptosis, Autophagy, and Mitophagy in the Loss of Plating Efficiency Induced by Bax Expression in Yeast", Journal of Biological Chemistry, vol. 281, pp. 36187-36197, 2006. http://dx.doi.org/10.1074/jbc.M607444200
- M. Ligr, F. Madeo, E. Fröhlich, W. Hilt, K. Fröhlich, and D.H. Wolf, "Mammalian Bax triggers apoptotic changes in yeast", FEBS Letters, vol. 438, pp. 61-65, 1998. http://dx.doi.org/10.1016/s0014-5793(98)01227-7
- C.M. Khoury, and M.T. Greenwood, "The pleiotropic effects of heterologous Bax expression in yeast", Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, vol. 1783, pp. 1449-1465, 2008. http://dx.doi.org/10.1016/j.bbamcr.2007.12.013
- R.D. Silva, S. Manon, J. Gonçalves, L. Saraiva, and M. Côrte-Real, "Modulation of Bax mitochondrial insertion and induced cell death in yeast by mammalian protein kinase Cα", Experimental Cell Research, vol. 317, pp. 781-790, 2011. http://dx.doi.org/10.1016/j.yexcr.2010.12.001
- G. Laloux, M. Deghelt, M. de Barsy, J. Letesson, and X. De Bolle, "Identification of the Essential Brucella melitensis Porin Omp2b as a Suppressor of Bax-Induced Cell Death in Yeast in a Genome-Wide Screening", PLoS ONE, vol. 5, pp. e13274, 2010. http://dx.doi.org/10.1371/journal.pone.0013274
- V. Ganesan, M.N. Perera, D. Colombini, D. Datskovskiy, K. Chadha, and M. Colombini, "Ceramide and activated Bax act synergistically to permeabilize the mitochondrial outer membrane", Apoptosis, vol. 15, pp. 553-562, 2010. http://dx.doi.org/10.1007/s10495-009-0449-0
- D. Williams, G. Norman, C. Khoury, N. Metcalfe, J. Briard, A. Laporte, S. Sheibani, L. Portt, C.A. Mandato, and M.T. Greenwood, "Evidence for a second messenger function of dUTP during Bax mediated apoptosis of yeast and mammalian cells", Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, vol. 1813, pp. 315-321, 2011. http://dx.doi.org/10.1016/j.bbamcr.2010.11.021
- E.L. Chua, L. Young, W.M. Wu, J.R. Turtle, and Q. Dong, "Cloning of TC-1 (C8orf4), a Novel Gene Found to Be Overexpressed in Thyroid Cancer", Genomics, vol. 69, pp. 342-347, 2000. http://dx.doi.org/10.1006/geno.2000.6348
- J. Park, Y. Jung, J. Kim, K. Kim, S. Ahn, K. Song, and I. Lee, "TC1 (C8orf4) is upregulated by cellular stress and mediates heat shock response", Biochemical and Biophysical Research Communications, vol. 360, pp. 447-452, 2007. http://dx.doi.org/10.1016/j.bbrc.2007.06.077
- M. Sunde, K.C.Y. McGrath, L. Young, J.M. Matthews, E.L. Chua, J.P. Mackay, and A.K. Death, "TC-1 Is a Novel Tumorigenic and Natively Disordered Protein Associated with Thyroid Cancer", Cancer Research, vol. 64, pp. 2766-2773, 2004. http://dx.doi.org/10.1158/0008-5472.can-03-2093
- J. Lei, W. Li, Y. Yang, Q. Lu, N. Zhang, G. Bai, D. Zhong, K. Su, B. Liu, X. Li, Y. Wang, and X. Wang, "TC-1 Overexpression Promotes Cell Proliferation in Human Non-Small Cell Lung Cancer that Can Be Inhibited by PD173074", PLoS ONE, vol. 9, pp. e100075, 2014. http://dx.doi.org/10.1371/journal.pone.0100075
- Z. Yang, C. Khoury, G. Jean-Baptiste, and M.T. Greenwood, "Identification of mouse sphingomyelin synthase 1 as a suppressor of Bax-mediated cell death in yeast", FEMS Yeast Research, vol. 6, pp. 751-762, 2006. http://dx.doi.org/10.1111/j.1567-1364.2006.00052.x
- C. Clapp, L. Portt, C. Khoury, S. Sheibani, G. Norman, P. Ebner, R. Eid, H. Vali, C.A. Mandato, F. Madeo, and M.T. Greenwood, "14-3-3 Protects against stress-induced apoptosis", Cell Death & Disease, vol. 3, pp. e348-e348, 2012. http://dx.doi.org/10.1038/cddis.2012.90
- C. Notredame, D.G. Higgins, and J. Heringa, "T-coffee: a novel method for fast and accurate multiple sequence alignment 1 1Edited by J. Thornton", Journal of Molecular Biology, vol. 302, pp. 205-217, 2000. http://dx.doi.org/10.1006/jmbi.2000.4042
- M. Nicod, S. Michlig, M. Flahaut, M. Salinas, N.F. Jaeger, J. Horisberger, B.C. Rossier, and D. Firsov, "A novel vasopressin-induced transcript promotes MAP kinase activation and ENaC downregulation", The EMBO Journal, vol. 21, pp. 5109-5117, 2002. http://dx.doi.org/10.1093/emboj/cdf509
- J. Cracraft, and M. Donoghue, "Assembling the tree of life. In: Cracraft J, Donoghue MJ, editors. Assembling the tree of life", Oxford University Press, New York; pp. 187., 2004.
- K. Miura, W. Fujibuchi, and M. Unno, "Splice variants in apoptotic pathway.", Experimental oncology, 2012. http://www.ncbi.nlm.nih.gov/pubmed/23070006
- C.M. Khoury, Z. Yang, X.Y. Li, M. Vignali, S. Fields, and M.T. Greenwood, "A TSC22-like motif defines a novel antiapoptotic protein family", FEMS Yeast Research, vol. 8, pp. 540-563, 2008. http://dx.doi.org/10.1111/j.1567-1364.2008.00367.x
- R. Eid, S. Sheibani, N. Gharib, J.F. Lapointe, A. Horowitz, H. Vali, C.A. Mandato, and M.T. Greenwood, "Human ribosomal protein L9 is a Bax suppressor that promotes cell survival in yeast", FEMS Yeast Research, vol. 14, pp. 495-507, 2013. http://dx.doi.org/10.1111/1567-1364.12121
- Z. Yang, A.B. Moffa, R. Haddad, K.L. Streicher, and S.P. Ethier, "Transforming properties of TC‐1 in human breast cancer: Interaction with FGFR2 and β‐catenin signaling pathways", International Journal of Cancer, vol. 121, pp. 1265-1273, 2007. http://dx.doi.org/10.1002/ijc.22831
- H. Xu, Y. Liu, S. Liu, Y. Miao, Q. Li, and E. Wang, "TC-1 (C8orf4) expression is correlated with differentiation in ovarian carcinomas and might distinguish metastatic ovarian from metastatic colorectal carcinomas", Virchows Archiv, vol. 462, pp. 281-287, 2013. http://dx.doi.org/10.1007/s00428-013-1375-7
- K. Su, L. Huang, W. Li, X. Yan, X. Li, Z. Zhang, F. Jin, J. Lei, G. Ba, B. Liu, X. Wang, and Y. Wang, "TC-1 (c8orf4) enhances aggressive biologic behavior in lung cancer through the Wnt/β-catenin pathway", Journal of Surgical Research, vol. 185, pp. 255-263, 2013. http://dx.doi.org/10.1016/j.jss.2013.05.075
- M. Ždralević, N. Guaragnella, L. Antonacci, E. Marra, and S. Giannattasio, "Yeast as a Tool to Study Signaling Pathways in Mitochondrial Stress Response and Cytoprotection", The Scientific World Journal, vol. 2012, pp. 1-10, 2012. http://dx.doi.org/10.1100/2012/912147
- S. Büttner, T. Eisenberg, D. Carmona-Gutierrez, D. Ruli, H. Knauer, C. Ruckenstuhl, C. Sigrist, S. Wissing, M. Kollroser, K. Fröhlich, S. Sigrist, and F. Madeo, "Endonuclease G Regulates Budding Yeast Life and Death", Molecular Cell, vol. 25, pp. 233-246, 2007. http://dx.doi.org/10.1016/j.molcel.2006.12.021
- A. Horowitz, J.F. Lapointe, R. Eid, S. Sheibani, N. Gharib, N.K. Jones, H. Vali, C.A. Mandato, and M.T. Greenwood, "The human septin7 and the yeast CDC10 septin prevent Bax and copper mediated cell death in yeast", Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, vol. 1833, pp. 3186-3194, 2013. http://dx.doi.org/10.1016/j.bbamcr.2013.09.004
- Y. Jung, S. Bang, K. Choi, E. Kim, Y. Kim, J. Kim, J. Park, H. Koo, R.T. Moon, K. Song, and I. Lee, "TC1 (C8orf4) Enhances the Wnt/β-Catenin Pathway by Relieving Antagonistic Activity of Chibby", Cancer Research, vol. 66, pp. 723-728, 2006. http://dx.doi.org/10.1158/0008-5472.CAN-05-3124
- D. Dirnberger, and K. Seuwen, "Signaling of Human Frizzled Receptors to the Mating Pathway in Yeast", PLoS ONE, vol. 2, pp. e954, 2007. http://dx.doi.org/10.1371/journal.pone.0000954
ACKNOWLEDGMENTS
We thank F. Madeo for plasmids. This work was supported by grants from NSERC and the Canadian Defense Academy to MTG. RW is the recipient of scholarship from Ontario’s Ministry of Research and Innovation.
COPYRIGHT
© 2015

Human Thyroid Cancer-1 (TC-1) is a vertebrate specific oncogenic protein that protects against copper and pro-apoptotic genes in yeast by Natalie K. Jones et al. is licensed under a Creative Commons Attribution 4.0 International License.