Back to article: Improvement of biochemical methods of polyP quantification
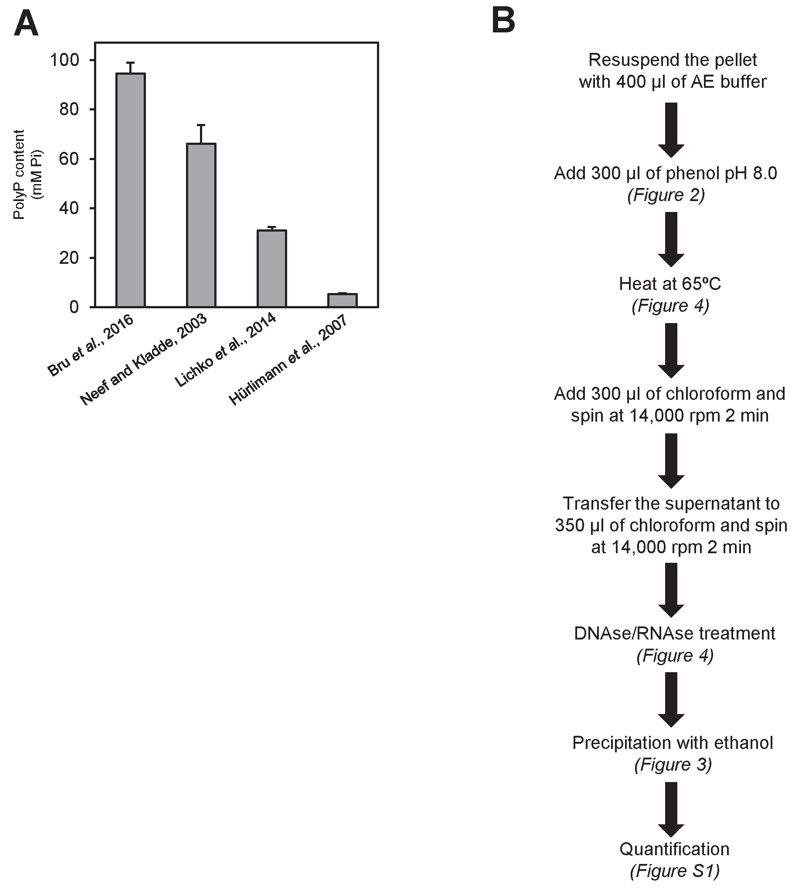
FIGURE 5: Comparison of different polyP extraction and purification methods.
(A) Amount of polyP corresponding to 107 logarithmically growing yeast cells extracted and purified with the following methods: neutral-phenol/chloroform and ethanol precipitation [26], acid-phenol/chloroform and ethanol precipitation [33], perchloric acid [23], and sulfuric acid and affinity columns [25]. Mean ± SEM from 3 independent experiments is shown.
(B) Scheme of the polyP extraction and purification protocol using neutral phenol and ethanol purification. In brackets appears the figure supporting this particular step. For details and tips see Materials and Methods.
23. Lichko LP, Eldarov MA, Dumina MV, and Kulakovskaya TV (2014). PPX1 gene overexpression has no influence on polyphosphates in Saccharomyces cerevisiae. Biochemistry (Mosc). 79(11): 1211-1215. https://doi.org/10.1134/S000629791411008X
25. Hurlimann HC, Stadler-Waibel M, Werner TP, and Freimoser FM (2007). Pho91 Is a vacuolar phosphate transporter that regulates phosphate and polyphosphate metabolism in Saccharomyces cerevisiae. Mol.Biol.Cell. 18(11): 4438-4445. https://doi.org/10.1091/mbc.e07-05-0457
26. Bru S, Martinez JM, Hernandez-Ortega S, Quandt E, Torres-Torronteras J, Marti RR, Canadell D, Arino J, Sharma S, Jimenez J, and Clotet J (2016). Polyphosphate is involved in cell cycle progression and genomic stability in Saccharomyces cerevisiae. Mol.Microbiol. 101(3): 367-80.https://doi.org/10.1111/mmi.13396
33. Neef DW, and Kladde MP (2003). Polyphosphate loss promotes SNF/SWI- and Gcn5-dependent mitotic induction of PHO5. Mol.Cell.Biol. 23(11): 3788-3797. https://doi.org/10.1128/MCB.23.11.3788-3797.2003