Back to article: Integrative metabolomics as emerging tool to study autophagy regulation
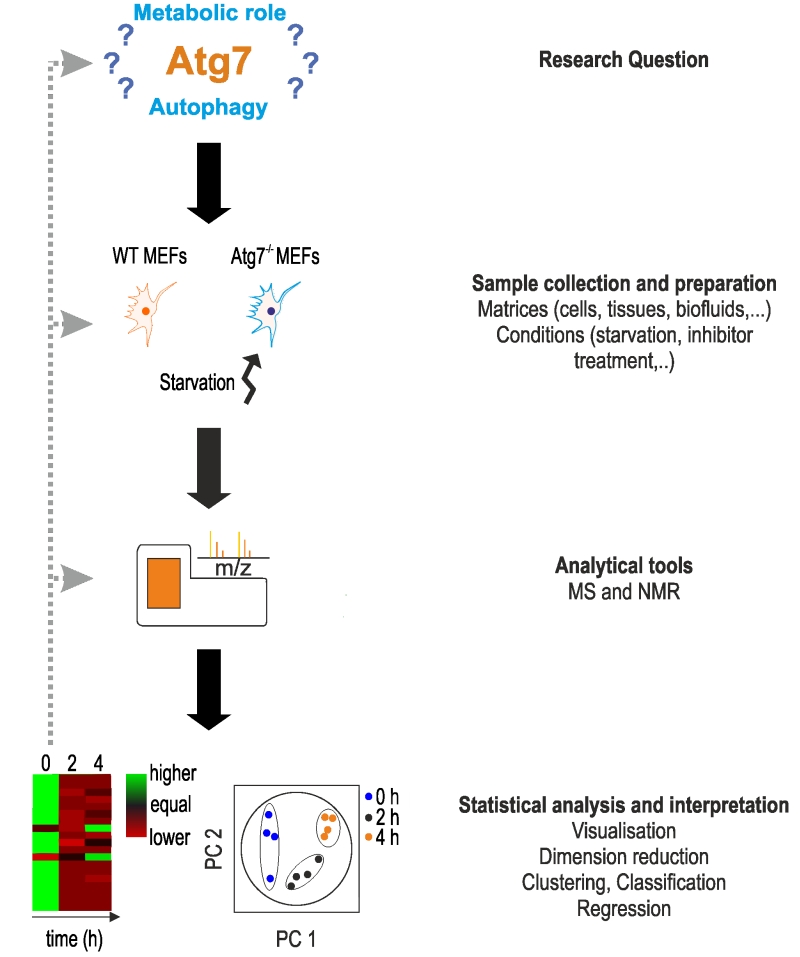
FIGURE 1: Characteristic setup of a metabolomics study. The setup of a metabo-lomics study is shown using the work of Shen et al. as an example [52]. Main steps are shown on the right. The main aim of the authors was to determine the function of Atg7, a protein essential in the formation of autophagosomes. In ab-sence of Atg7, cells indicate a reduction or lack of autophagy, however, the metabolic responses under acute starvation re-mained elusive. The main aim of the study was to determine the metabolic phenotype of Atg7-dependent autophagy under star-vation. Based on this research question, the authors used wild-type and Atg7-/- mouse embryonic fibroblasts (MEFs) as a model system and starvation as experimental condi-tions. Examples for other material and conditions can be found on the right. After treatment, cells were lysed for metabolomics analysis on an UPLC-MS system. Apart from MS analysis, NMR spectroscopy can be applied. Metabolomics data were analyzed statistically using principle component analysis (PCA), and heat maps were used to visualize changes in metabolite concentra-tions. For more details, see text.
52. Shen S, Weng R, Li L, Xu X, Bai Y, Liu H (2016). Metabolomic Analysis of Mouse Em-bryonic Fibroblast Cells in Response to Autophagy In-duced by Acute Starvation. Scientific reports 6:34075. https://doi.org/10.1038/srep34075