Reviews:
Microbial Cell, Vol. 6, No. 6, pp. 267 - 285; doi: 10.15698/mic2019.06.679
Yeast AP-1 like transcription factors (Yap) and stress response: a current overview
1 Instituto de Tecnologia Química e Biológica Anónio Xavier, Universidade Nova de Lisboa, Avenida da República, EAN, Oeiras 2781-901, Oeiras, Portugal.
2 Sorbonne Université, CNRS, Institut de Biologie Paris-Seine, Laboratory of Computational and Quantitative Biology, F-75005, Paris, France.
Keywords: yeast, Yap factors, cis-elements, bZIP, stress.
Abbreviations:
ARE – AP1-recogniction element,
bZIP – basic leucine-zipper,
CBC – CCAAT binding complex,
CIA – cytosolic iron–sulfur protein assembly,
CRD – cysteine-rich domain,
CWI – cell wall integrity,
ER – endoplasmic reticulum,
GSH – reduced glutathione,
GSSG – oxidized glutathione,
HOG - high osmolarity glycerol,
HQ – hydroquinone,
NES – nuclear export signal,
NO – nitric oxide,
ORF – open reading frame,
ROS – reactive oxygen species,
Trx – thioredoxin,
uORF – upstream ORF,
WGD- whole genome duplication,
Y8RE – Yap8 response element,
YRE – Yap response elements.
Received originally: 24/01/2019 Received in revised form: 23/04/2019
Accepted: 09/05/2019
Published: 28/05/2019
Correspondence:
C. Rodrigues-Pousada, Instituto de Tecnologia Química e Biológica Anónio Xavier, Universidade Nova de Lisboa, Avenida da República, EAN, Oeiras 2781-901, Oeiras, Portugal; claudina@itqb.unl.pt
Conflict of interest statement: The authors have no conflict of interest to declare.
Please cite this article as: Claudina Rodrigues-Pousada, Frédéric Devaux, Soraia M. Caetano, Catarina Pimentel, Sofia da Silva, Ana Carolina Cordeiro and Cata-rina Amaral (2019). Yeast AP-1 like transcription factors (Yap) and stress response: a current overview. Microbial Cell 6(6): 267-285. doi: 10.15698/mic2019.06.679
Abstract
Yeast adaptation to stress has been extensively studied. It involves large reprogramming of genome expression operated by many, more or less specific, transcription factors. Here, we review our current knowledge on the function of the eight Yap transcription factors (Yap1 to Yap8) in Saccharomyces cerevisiae, which were shown to be involved in various stress responses. More precisely, Yap1 is activated under oxidative stress, Yap2/Cad1 under cadmium, Yap4/Cin5 and Yap6 under osmotic shock, Yap5 under iron overload and Yap8/Arr1 by arsenic compounds. Yap3 and Yap7 seem to be involved in hydroquinone and nitrosative stresses, respectively. The data presented in this article illustrate how much knowledge on the function of these Yap transcription factors is advanced. The evolution of the Yap family and its roles in various pathogenic and non-pathogenic fungal species is discussed in the last section.
INTRODUCTION
The yeast Saccharomyces cerevisiae has been used in research for more than one hundred years, and it is generally regarded as the most well understood eukaryotic organism in the stress response field. The sensing and transduction of the stress signals into different cellular compartments induce a genetic reprogramming, which leads to a transient arrest of normal cellular processes, with a decrease in the expression of housekeeping genes and protein synthesis. In addition, there is an induction of the expression of genes encoding stress proteins such as molecular chaperones responsible for maintaining protein folding [1]. Survival and growth resumption imply successful cellular adaptation to the new conditions as well as the repair of damages incurred to the cell that might compromise its viability. Specific stress conditions elicit distinct cellular responses due to gene expression programs orchestrated by a number of specific transcription factors commonly activated when the cells shift to sub-optimal growth conditions. Among these transcription factors, the basic leucine-zipper (bZIP) proteins form a large multifunctional family, which is conserved in all eukaryotes [2]. These regulators play important roles in the maintenance of cellular homeostasis and in cell differentiation during development in multicellular organisms. They are defined by a basic DNA binding region followed by a leucine zipper motif. In metazoans, bZIP can form hetero-or homodimers, but yeast members of this family mostly act as homodimers [2]. Several subfamilies of bZIP regulators can be defined based on the protein sequences and DNA binding preferences [3]. In this review, we will highlight the role of the Yeast Activator (AP1-like) Protein (Yap) sub-family in the yeast adaptation to environmental stress response. The last section provides an overview of the evolution and functional significance of this family in other fungal species.
THE YAP FAMILIY OF TRANSCRIPTIONAL REGULATORS
Fifteen bZIP proteins are found in the S. cerevisiae genome. Four of them are homologous to the ATF/CREB subfamily (Aca1, Sko1, Hac1 and Cst6) and one is related to AP1 (Jun/Fos) transcription factors (Gcn4). The rest belongs to fungal specific bZIP subtypes [2]. The yeast activator (AP1) protein family is the largest bZIP subfamily in S. cerevisiae. It includes eight members (Yap1 to Yap8) which have some sequence similarity to Gcn4. Gcn4 interacts with DNA, via five residues in its basic region (Asn235, Ala238, Ala239, Ser242, and Arg243) that make base-specific contacts with DNA (Fig. 1). These residues are highly conserved in the Jun/Fos bZIP proteins found in mammals [4][5][6]. The Yap family is unusual among bZIP proteins because they contain a glutamine at the position corresponding to Gcn4 Ala239 and a phenylalanine or a tyrosine at the position corresponding to Ser242, hence having different DNA binding properties (Fig. 1). Furthermore, there are two family specific residues in the Yap family, in position 234 and 241 of Gcn4 that are a glutamine and an alanine, respectively [7].
–
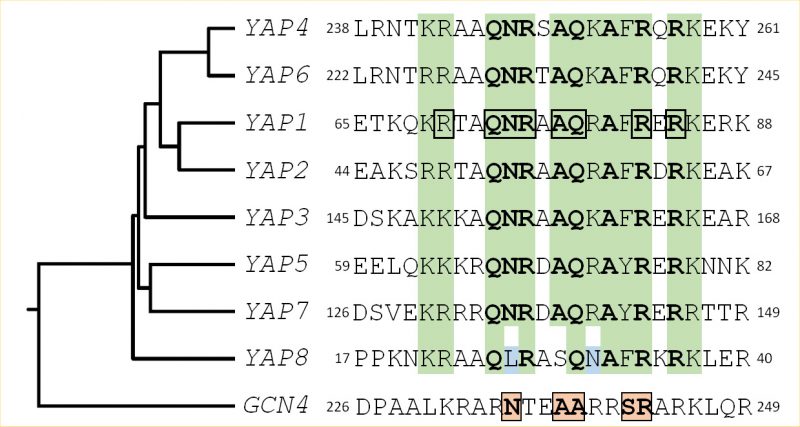 | FIGURE 1: Structural features of the Yap family DNA binding domain. The sequences of the eight Yap DNA binding domains (i.e. the basic region of the bZIP motif) are compared with the equivalent region of Gcn4, the classical yeast AP-1 factor, used as an outgroup. A green background highlights the positions, whose physico-chemical properties are conserved in the Yap family. The most conserved residues are in bold. The Yap8 specific residues are in blue. The Yap1 amino-acids which were predicted to contact DNA based on structural studies [12, 140] have been underlined by a black box. The Gcn4 residues involved in DNA interaction are highlighted by pink boxes. The rooted tree and the multiple alignment were obtained from ClustalW (https://www.genome.jp/tools-bin/clustalw), using the bZIP sequences and the 100 flanking amino-acids. |
–
Yap1, the first member of the Yap family to be described, was initially identified by its ability to bind a DNA sequence containing the simian virus 40 (SV-40) sequence AP-1 recognition element (ARE: TGACTAA). Based on the ARE-binding capacity, this factor was purified as a 90 kDa protein and the corresponding gene was cloned by screening a λgt11 library with a monoclonal antibody raised against Yap1 [8]. Subsequently, this gene was also found as a multicopy suppressor of sensitivity to the iron chelators 1,10-phenantroline as well as to a variety of drugs, including cycloheximide. Hence, this locus was historically designated as PAR1/SNQ3/PDR4 [9]. Besides YAP1, a second gene, YAP2, conferring resistance to the iron chelator 1,10-phenantroline, was also described. This gene encodes a 45 kDa protein that binds YRE (Yap response elements) located in the promoters of its targets. YAP2 is also named CAD1, due to the acquisition of cadmium resistance in cells overexpressing this gene [10]. The sequencing of YAP1 and YAP2 genes revealed the presence of three conserved regions: the bZIP domain in the N-terminus, a region in the C-terminus containing conserved cysteine residues and another one in the internal region adjacent to the bZIP-domain [7].
–
A search in the S. cerevisiae genome using as query the bZIP motif revealed the other six members of the Yap family [11]. All of them possess common key residues in the bZIP, which confer to the family distinct DNA binding properties (Fig. 1).
–
Yap1 recognizes the specific sequences TGACTAA, TTAGTCA, TTACTAA and T(T/G)ACAAA (YREs) in the promoter of its target genes [11][12][13][14]. Genome-wide analyses have defined the consensus Yap1 sequence as being TTACTAA (YRE-O) [12][15][16]. The remaining Yap transcription factors bind either the YRE-O element (Yap2/Cad1, Yap5, Yap7) or a slightly different motif, TTACGTAA, called YRE-A (Yap4/Cin5, Yap6) [16][17][18][19]. Yap3 was described as a transactivator of the YRE-O, but the YRE-A was predicted as his preferred binding motif based on chromatin immuno-precipitation (ChIP-chip) experiments [11][12][13][14][15][16][17]. The preference for YRE-O or YRE-A has been proposed to be due to the presence of either an arginine or a lysine in the basic domain of the corresponding Yap (position 15 in the sequences represented in Fig. 1) [17], however, this hypothesis is controversial [11][12]. The sole exception is Yap8/Arr1, which binds a cis-element with 13 base pair sequence TGATTAATAATCA hereafter designated as Yap8 response element (Y8RE) [20][21]. Both the core element (TTAATAA) and the flanking regions (TGA and TCA) of Y8RE are crucial for Yap8/Arr1 binding and for in vivo activation of its targets [20][21]. Interestingly, a residue in the Yap8 basic region, Leu26, is required for Yap8-DNA binding and Yap8 activity (highlighted in blue in Fig. 1). This residue, together with Asn31, hinders Yap1 response element recognition by Yap8, giving its narrow DNA-binding specificity [20].
–
A structural common feature between YAP1 and YAP2 is the presence of unusually long 5'-untranslated region containing short upstream open reading frames (uORF). The YAP1 leader has one 7-codon uORF whereas the one of YAP2 contains one 6-codon uORF (uORF1) and an overlapping short reading frame of 23 codons (uORF2), which is located at -1 with respect to the main reading frame [22]. The latter is involved in YAP2 mRNA turnover via termination-dependent decay. Indeed, the YAP1-type uORF allows scanning of 40S subunits to proceed via leaky scanning and re-initiation to the major ORF, whereas the YAP2-type acts to block ribosomal scanning by promoting efficient termination [23].
YAP1, THE REGULATOR OF OXIDATVE STRESS RESPONSE
Cells have to keep intracellular concentrations of peroxides (H2O2 and organic peroxides) and of other reactive oxygen species (ROS) at very low levels by regulating their concentration through tightly controlled mechanisms. ROS are endogenously produced during aerobic respiration or because of altered cellular environment (oxidant molecules exposure, imbalance in metal homeostasis). Microorganisms, including S. cerevisiae, contain sensors that detect the levels of ROS. In yeast cells, Yap1 is activated under oxidative stress conditions and its absence renders cells hypersensitive to the several oxidants that generate superoxide anion radicals [9]. Kuge and Jones provided the first and clear clue towards the role of Yap1 in this response mechanism [24], by showing that TRX2 gene (thioredoxin) induction by H2O2, t-BOOH, diamide and diethylmaleate (DEM) is Yap1-dependent. Analyses of several promoter sequences of antioxidants genes such as TRX2, GSH1 (γ-glutamyl cysteine synthetase) [24], GSH2 (gluthathione synthetase) [25] and TRR1 (thioredoxin reductase) [26] revealed functional YREs.
–
Yap1 redox regulation by oxidants involves two cysteine-rich domains (CRDs) located in the N- and C-terminus. Yap1 shuttles between the nucleus and the cytoplasm mediated by the exportin Crm1/Xpo1 and imported by the importin Pse1 [27][28]. Upon stress conditions, Yap1 accumulates in the nucleus because its NES (nuclear export signal) is masked by the formation of an intramolecular disulfide bond between the cysteine 303 and 598, avoiding Crm1 recognition. Consequently, there is an increased expression of Yap1 targets (Fig. 2) [28]. In vitro studies performed by Wood M.J. et al. revealed that upon H2O2 exposure, an additional intramolecular disulfide bond between Cys310 in n-CRD and Cys629 in c-CRD is formed [29]. Although this second disulfide bond was not shown to be relevant in vivo, it possibly adds stability to the oxidized active conformation of Yap1 [30]. The fact that Yap1 does not respond to H2O2 in the absence of Hyr1 (also designated as Gpx3 or Orp1), led to establish the role of this protein as a sensor and signal transducer of H2O2. Yap1 oxidation does not thus take place directly and the Cys36 of Hyr1/Orp1 oxidized to sulfenic acid (Cys36-SOH) senses the H2O2 signal [31][32]. Next, the signal is transduced to Yap1 through the generation of an intermolecular disulfide bond between Cys36 of Hyr1/Orp1 and Yap1 Cys598, which then forms the intramolecular disulfide bridge with Yap1 Cys303 rendering Yap1 to its active form (Fig. 2) [29]. When this bond is formed, the Cys36 sulfenic acid of Hyr1/Orp1 is prepared to react with its Cys82 to complete the peroxidative cycle. Bersweiler et al. showed that the protein Ybp1 could be associated to Yap1 forming a ternary complex with Hyr1/Orp1 [33]. It is possible that Ybp1 functions as chaperoning the formation of the disulfide bridge between the Cys36-SOH of Hyr1/Orp1 with the Cys598 of Yap1. It could also avoid the competition with the Cys36-Cys82 disulfide bond of Hyr1/Orp1 that is part of its catalytic site [33]. The Hyr1/Orp1 peroxidase is different from the classical ones and is reduced by the thioredoxin (Trx) pathway [32][34].
–
–
In contrast to H2O2, the Yap1 response to diamide is Hyr1/Orp1-independent and it does not involve the n-CRD (Fig. 2) [35]. This is consistent with the notion that it possesses two redox centers. Indeed, the electrophile N-ethylmaleimide (NEM) and the quinone menadione, both electrophile and superoxide anion generators, were shown to modify the c-CRD cysteines independently of Hyr1/Orp1, leading to Yap1 nuclear accumulation [35]. The Trx pathway is involved in the recycling of the Yap1 oxidized form by disrupting the disulfide bond [36][37][38]. Fe-S clusters are also very susceptible to oxidation. It was described that Yap1 attenuates the toxic effect of hydroxyurea by regulating the expression of key players of the cytosolic iron–sulfur protein assembly machinery (CIA), proposed to act as sensors of the intracellular oxidative stress [39][40].
–
During the oxidation-reduction processes in which Yap1 is active, the glutathione functions either in reduced (GSH) or oxidized (GSSG) form. Recently, it was shown that glutathione in the endoplasmic reticulum (ER) is oxidized but not reduced, being catalyzed to the oxidative state by Ero1, a protein forming the disulfide bond necessary for this process. The reduction of this GSSG molecule to GSH is then occurring in the cytoplasm. As such, the interplay between reduced cytosolic GSH and the oxidized GSSG in the ER keeps the redox homeostasis [41][42]. The transport of glutathione between the cytoplasm and the ER is facilitated via diffusion through the Sec61 complex (protein-conducting channel) plus the Sec62-Sec63 complex [41][42].
YAP1 IN METAL AND METALLOID STRESS
Metal toxicity depends on each metal's physicochemical properties and ligand preferences. Redox-active metals such as iron (Fe), chromium (Cr), copper (Cu) and cobalt (Co) can take oxygen and sulphur as their ligands, whereas redox-inactive metals such as cadmium (Cd) and mercury (Hg) prefer sulphur as a ligand [43][44][45]. Redox-active metals can induce oxidative stress by participating in Fenton-type reactions, whereas redox-inactive metals imbalance the antioxidant pool of the cell [43][44][45]. In general, metals induce cellular toxicity by generating oxidative stress, impairing the DNA repair system and inhibiting protein folding and function [45][46][47].
–
Yap1 plays a pivotal role in mitigating metal-generated ROS, but its contribution to metal detoxification is not restricted to the induction of the cellular antioxidant defences. Indeed, Yap1 also regulates expression of the YCF1 gene, the vacuolar transporter of metal-glutathione conjugates, thereby contributing to vacuolar compartmentalization of metals and metalloids such as cadmium and arsenite [48][49]. Over the last years, several other metal detoxification pathways controlled by Yap1 were unveiled as subsequently detailed.
–
Cobalt is a biologically relevant metal in many living organisms because it is an essential cofactor of enzymes involved in many reactions [50]. However, when cobalt is in excess it generates oxidative stress, which damages cells. Analysis of transcriptional profiles of cells stressed with high concentrations of cobalt revealed the induction of antioxidant genes in a Yap1 dependent way [51]. Corroborating this molecular data, biochemical analysis showed Yap1 to be important to deal with oxidative damage generated by exposure to cobalt. Activation of Yap1 is not exclusively involved in cobalt-generated ROS since under anoxia, Yap1 also localizes in the nucleus [51]. Moreover, Yap1 up-regulates cobalt uptake through the activation of the expression of the high affinity phosphate transporter PHO84, a well-known cobalt transporter. Accordingly, yap1 knockout cells accumulate lower levels of cobalt [51]. The authors suggested that cobalt accumulation could be a side effect of Yap1 regulation of PHO84 under non-stressed conditions and proposed that phosphate uptake mediated by Yap1 may fulfill a role in the oxidative stress response triggered by the aerobic metabolism. Reinforcing this possibility, growing evidences indicate that manganesephosphate complexes, which enter cells via Pho84 [52], act as scavengers of superoxide [53][54].
–
Cadmium is a well-known mutagenic metal that can enter cells via non-specific divalent metal transporters. Yap1 is a repressor of the FET4 gene [51], a plasma membrane low affinity iron transporter, which can transport other bivalent metals including cobalt and cadmium ions. Although this repression does not significantly affect cobalt uptake, it avoids cadmium toxicity by impairing its transport into the cell [55]. Genomic deletion of Yap1 increases FET4 transcripts as well as protein levels [55]. The yap1 mutant accumulates high cadmium levels compared with the wild-type strain, whilst the deletion of FET4 gene from the yap1 mutant resumes cadmium tolerance. Noteworthy, cadmium uptake increased in cells treated with both cadmium and iron because iron induces CUP1 expression, which possibly binds and sequesters cadmium [55]. Yap1 is not a direct regulator of FET4 because its promoter does not contain YREs. Previous microarray analysis obtained in the presence of cobalt [51] revealed that Yap1 positively regulates the transcription factor Rox1. This factor is a repressor of hypoxic genes and represses FET4 expression under aerobic conditions. The promoter of ROX1 possesses one functional YRE located at – 414 upstream the ATG codon. Yap1 is a direct regulator of ROX1, which in turn represses FET4 [55].
–
Yap1 also plays an important role in arsenic compound detoxification by regulating genes encoding several of the cellular antioxidant defenses, important to mitigate arsenic-generated ROS [56]. Besides, Yap1 was also shown to control the expression of YCF1, ACR2, the yeast arsenate reductase gene, and ACR3, the plasma membrane arsenite-efflux protein-encoding gene [48][57][58]. Recently, a new line of action of Yap1 in the protection against arsenate toxicity was put forward. By analyzing the transcriptomic profile of Yap1 knockout cells treated with arsenate, several genes involved in the biogenesis of mitochondrial (ISC) and cytosolic (CIA) Fe-S clusters were found to be dependent on Yap1 [59]. This dependence was maintained under anoxia, suggesting that arsenate per se is able to activate Yap1 and triggers the up-regulation of Fe-S cluster biogenesis genes. Arsenate was shown to directly and indirectly (possibly via intracellular ROS production) affect the activity of Fe-S containing proteins and accordingly overexpression of CIA and ISC genes attenuates arsenate deleterious effects [59]. Together these findings led the authors to propose that the transcriptional regulation of Fe-S biogenesis genes may constitute another safeguard against arsenate toxicity activated by Yap1.
YAP2/CAD1 INVOLVEMENT IN CADMIUM AND OXIDATVE STRESS
Yap2/Cad1 is the family member that shares the highest homology with Yap1 [10]. When overexpressed, Yap2 confers resistance to several stress agents, suggesting a role for this transcription factor in response to toxic compounds. Although YAP2 and YAP1 overexpression elicits similar phenotypes, deletion of the latter has strong phenotypic effects, whereas deletion of YAP2 does not affect or only slightly affects cell growth [7][10].
–
Notably, the YAP2 and YAP1 single deletion similarly decreased the resistance towards the oxidants H2O2 and menadione of stationary-phase cultures [60]. Under such circumstances, Yap2 does not regulate the known Yap1 antioxidant targets, an observation that led the authors to propose that the H2O2-mediated adaptive response could be composed of two distinct regulons, one being controlled by Yap1 and the other by Yap2 [60]. These data also support the notion that Yap1 and Yap2 have overlapping, but not redundant functions. Corroborating this idea, the analysis of the transcriptomic profile of yap1 and yap2 null mutants showed that Yap1 and Yap2 activate separated regulons when challenged with H2O2 [61].
–
Yap2 transactivation potential is slightly stimulated upon treatment with cadmium [11]. The swapping of Yap1 and Yap2 c-CRDs domains shows that Yap2 cCRD can function in the context of Yap1 in response to cadmium but not in response to H2O2, indicating the high specificity of these responses [62]. Accordingly, overexpression of YAP2 in the yap1 null mutant suppresses cadmium, but not H2O2 sensitivity [63].
–
Yap2 is mainly localized in the cytoplasm in unstressed cells but soon after the addition of cadmium it accumulates in the nucleus, activating its targets. In the Yap2 C-terminus, there are three cysteines, Cys391, Cys356 and Cys387, to which cadmium binds directly as shown using the high molecular mass alkylating agent (AMS), which targets free thiol moieties in cysteines. This interaction masks the nuclear export signal recognized by the exportin Crm1 leading to the accumulation of Yap2 in the nucleus [62].
–
Using proteomic analysis after cadmium treatment, Azevedo et al. searched for other Yap2-specific targets [62]. In order to eliminate the influence of Yap1-target genes, a yap1 null strain transformed with a Yap2 multi-copy vector was used in the presence or absence of cadmium. Proteome analysis under such conditions revealed the induction of the Frm2 protein. The expression of the gene encoding this protein is only dependent on Yap2 in the presence of cadmium. Frm2 shares high identity with type 4 nitrore-ductases, shown to be involved in the fatty acid signalling pathway and required for unsaturated fatty acid control of the stearoyl-CoA desaturase gene (OLE1) expression [64]. FRM2 was also identified in a screen for mutants defective in OLE1 repression by unsaturated fatty acids, and the fact that the frm2 mutant is sensitive to arachidonic acid led to the hypothesis that FRM2 participates in lipid metabolism [62]. Considering that cadmium exerts its toxicity by promoting lipid peroxidation cascades, it is plausible that Yap2 regulates lipid metabolism [65][66].
–
Yap2 was also found in a two-hybrid screen using Rck1 or Rck2 MAPK-activated protein kinases (MAPKAPs) as baits [67][68]. The sensitivity of the rck1 mutant to tBOOH is fully suppressed by overexpression of Yap2 [67]. Although Yap2 is a cadmium responsive transcription factor, its deletion does not increase yeast sensitivity to cadmium. In the absence of Rck1, Yap2 gives protection against cadmium toxicity. These results indicate that Rck1 appears to have an inhibitory effect on Yap2 activity. Furthermore, Yap2 may play a role in cell wall maintenance by controlling the expression of CWI (Cell wall Integrity) genes, namely SLT2, RLM1 and CHS1 [68][69]. These genes are dependent on Yap2 but not on Yap1 and in the yap2 mutant strains, CWI genes are downregulated in the presence of cadmium indicating a regulatory role of Yap2 on their expression. It is possible that cadmium causes damage in the glucan structure of the cell wall, thus activating the expression of the CWI genes [68][69].
YAP3, A TRANSCRIPTION FACTOR WITH A POTENTIAL FUNCTION UNDER HYDROQUINONE STRESS
Yap3 (YHL009C) encodes a 399 amino acid protein containing a bZip domain similar to the other family members (Fig. 1). YAP3 is located in the chromosome VIII and activates transcription from promoters containing a Yap recognition element (YRE; 5'-TTAC/GTAA-3') [11]. YAP3 is not an essential gene and so far, the regulatory targets of Yap3 are not yet defined but it seems that Yap3 plays a specific role in the cellular response to hydroquinone (HQ). Indeed, the yap3 mutant strain is sensitive to HQ. Like other Yap family members, Yap3 contains two CRDs and a NES in its C-terminus. Yap3 localizes in the nucleus upon treatment with HQ [70]. Yap3 also responds to ER stress, as the null mutants are sensitive to tunicamycin, a compound that causes ER stress through induction of the unfolded protein response [71]. Yap3 was also identified in a screen of wild-type and mutant strains as being sensitive to arsenic (As) and monomethylarsonous (MMA) treatments, suggesting that these stresses and HQ share cellular targets [72].
–
Yap3 possesses a very high transactivation potential, even higher than the one of Yap1 in the absence of any stress [11], suggesting that this transcription factor might have an important function which was not yet precisely identified.
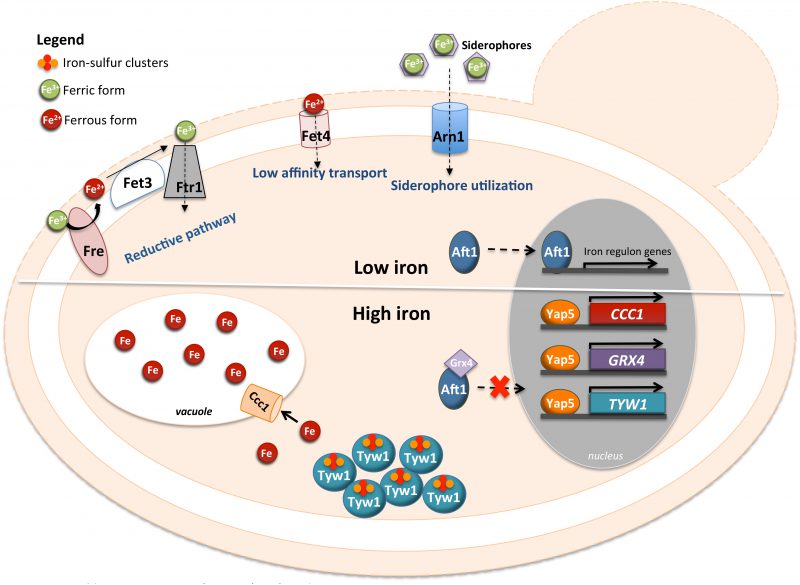
YAP5 CONTRIBUTES TO IRON HOMEOSTASIS
Yap5 (YIR018W) is a protein containing a CRD at the C-terminus and a bZIP domain at the N-terminus that recognizes YRE-O sites (Fig. 1). Additionally, Yap5 possesses a Hap4L domain just upstream of its bZIP. The Hap4L motif is a conserved protein sequence of 16 amino acids, which is found in proteins interacting with the CCAAT binding complex (CBC), a highly conserved transcriptional regulator [73]. However, the Hap4L domain of Yap5 is degenerated compared to other CBC interacting proteins and its role in Yap5 activity in S. cerevisiae has not been investigated yet [19]. Yap5 is responsible for yeast adaptation to iron overloading conditions. The Ccc1 transporter fulfills an important role in high iron detoxification, by importing iron into the vacuole, which is the major site of iron storage in fungi and plants [18]. CCC1 expression is induced by iron through the activity of Yap5 [18][74]. However, the up-regulation of CCC1 expression driven by Yap5 is not essential for cells to cope with high iron toxicity, since deletion of the functional YRE [18] from the CCC1 promoter region still allows cell growth under high iron levels [74]. Corroborating this notion, the yap5 null mutant is not as sensitive to iron excess as the ccc1 null mutant is [74][75][76].
–
Transcriptional and chromatin immunoprecipitation analyses revealed two other genes directly regulated by Yap5 with a relation to iron homeostasis, TYW1 and GRX4 [74][77][78]. TYW1 encodes a cytosolic iron-sulfur (Fe-S) cluster-containing enzyme required for the synthesis of Wybutosine modified tRNA [79]. It was proposed that the induction of TYW1 triggered by Yap5 might provide protection against iron toxicity by sequestering cytosolic free iron as protein-bound Fe-S clusters [77]. GRX4 encodes a cytosolic monothiol glutaredoxin, which together with Grx3 inhibits Aft1 activity under iron loading conditions by promoting its retention in the cytoplasm [80][81]. The Yap5-dependent up-regulation of GRX4 expression was suggested to reinforce this function, as in the yap5 null mutant Aft1 nuclear exclusion is slightly impaired [74] (Fig. 3).
–
–
While the transcriptional activity of Yap5 depends on iron bioavailability, Yap5 binding to the promoter of its target genes is iron insensitive [18][74] and Yap5 is constitutively localized in the nucleus. Iron sensing by this transcription factor depends on two CRDs (amino- and carboxyl-terminal CRDs), located in the C-terminus of the protein and separated by 37 residues [18]. Mutations of the cysteine residues of the CRDs impair the induction of Yap5 targets and compromise the adaptation of yeast to high iron [18].
–
Yap5 activation by iron is abrogated by mutations in genes encoding proteins of the mitochondrial iron-sulfur cluster assembly system (ISC), indicating that the transcriptional response to high iron is dependent on Fe-S biogenesis [82]. Accordingly, Rietzschel et al. showed that Yap5 senses iron by coordination of Fe-S clusters [76]. Both CRDs of Yap5 bind a 2Fe-2S cluster, whose maturation is uncommon as it depends on the ISC but not on the cytosolic CIA machinery [76]. These authors found that Fe-S cluster binding to Yap5 induces a conformational change in the protein, which may explain the increase in its transactivation potential under conditions of iron excess.
THE ROLE OF YAP4/CIN5 AND YAP6 DURING OSMOTIC STRESS
Hyperosmotic stress leads to the passive efflux of water from the cell to the exterior, resulting in a decrease in cell volume, loss of the state of turgidity, resulting in rigidity and increased concentration of cellular solutes [83]. In the case of hypo-osmotic environment, it allows the movement of water into the cell, originating the swelling of the cell, a high-pressure turgor as well as the dilution of the intracellular milieu [83]. To counteract these effects, the cell makes use of osmolytes, which are compatible solutes, such as the alcohol glycerol, trehalose, and sorbitol that protect the cell against the effects of an osmotic challenge by modifying the intracellular osmotic pressure [83][84][85]. These alterations correlate to modifications of gene expression that consequently leads to the alteration of the cell permeability to the osmolytes and of their biogenesis rate. These metabolic alterations are triggered by the HOG (high osmolarity glycerol) pathway via the modulation of the expression of stress-responsive genes [86]. The main actor of this pathway is Hog1, a mitogen-activated protein kinase (MAPK). Hog1 controls the activity of several transcription factors, in particular Msn1 [87], Msn2/4 [88], Hot1p [88], and Sko1p [86], among others.
–
Yap4/Cin5, the fourth member of the Yap family was initially characterized as a chromosome instability mutant designated as Cin5, encoding a 33 kDa protein [89]. Its overexpression confers salt tolerance [90] as well as resistance to antimalarial drugs [91] and cisplatin [92]. Results from several microarray analyses indicate an induction of Yap4 under various conditions, such as those of oxidative and osmotic stress. The YAP4 null mutant reveals a slight salt-sensitivity phenotype under hyperosmotic stress. On the other hand, it was also shown that under these conditions YAP4 gene expression is regulated by Msn2 through the more proximal STRE elements (-430bp). The promoter of YAP4 contains several cis-elements such as HSE (nGAAnnTTCn) located at -432 and -425 upstream of ATG codon and bound to HSF, a Yap1-cis element located at -516 and -508 from the ATG codon (YRE – TTAG/CTAA) and also STRE cis-elements for Msn2/Msn4 (AGGGG) [90]. Moreover, Yap4 is a downstream component of the HOG pathway and its overexpression partially rescues the salt sensitive phenotype of the hog1 single mutant [90].
–
Yap6 is a 44 kDa protein sharing almost 20% identity with Yap4/Cin5, making it the closest-related Yap family members (Fig. 1). Overexpression studies in the ena1 mutant (lacking the Na+/Li+ extrusion ATPase) subsequently identified both YAP4/CIN5 (HAL6) and YAP6 (HAL7) as genes that confer salt tolerance through a mechanism unrelated to the Na+/Li+ ATPase extrusion [93]. In contrast to Yap1, Yap2 and Yap8, the subcellular localization of Yap4 and Yap6 is constitutively nuclear.
–
Yap4 also interacts with the product of the yeast gene LOT6 (YLR011W) encoding a 21 kDa protein. Lot6 possesses a quinone reductase activity similar to its mammalian counterparts [90][94][95]. The association of Yap4 with this quinone reductase and the 20S proteasome affects its ubiquitin-independent degradation. It was proposed that the FMN cofactor in the Lot6 active site is a redox-regulated switch that controls the stability and localization of Yap4 [96]. A similar redox-controlled mechanism might regulate p53 and related transcription factors in mammalian cells [97]. It was also reported that the association of Lot6 with the 20S proteasome is via its flavin-binding site [96]. Interestingly, however, these authors showed that the reduction of the FMN cofactor by either NADH or light irradiation results in the binding of Yap4 to the Lot6–proteasome complex, indicating that recruitment of Yap4 depends on the redox state of the quinone reductase [96]. Alternatively, Lot6 in its native dimeric state is essential for the binding of Yap4 to the complex. The dissociation of Lot6 dimers into monomers does not affect the catalytic properties of the enzyme with regard to quinone reduction [98]. These authors put forward the hypothesis that Yap4 binds the Lot6:20S proteasome in a redox-dependent manner and may participate in the proapoptotic effect of Lot6 and thus might represent an activator of yeast apoptosis [98].
–
Transcriptional arrays of the yap4 mutant under mild conditions of hyperosmolarity revealed a large set of genes possibly regulated by Yap4. Amongst these target genes are GCY1, encoding a putative glycerol dehydrogenase, and GPP2, encoding a NAD-dependent glycerol-3-phosphate phosphatase. These genes show a decrease of their induction in the yap4 mutant strain with reduction values corresponding to 40% and 50% of the maximum levels, respectively. Furthermore, DCS2, a gene homologous to the DCS1-encoded mRNA decapping enzyme, shows 80% reduction of its induction level in the yap4 mutant upon osmotic shock. The fact that YAP4 and YAP6 are induced by a variety of unrelated forms of environmental stresses suggests a universal role in the yeast response to stress, in contrast to the other Yap members [90].
–
ChIP-chip experiments have shown that Yap4/Cin5, Sko1, Yap6, Msn2 and Skn7 bind their targets after incubation with high salt (0.6 M) for 30 min [99][100]. Later, Ni et al. determined that the binding of several of these transcription factors is a dynamic process [101]. Their data allowed the classification of Yap4 targets into three classes: constant binding independently of salt (class 1), rapid induction (class 2) and slow induction (class 3). Other minor binding patterns were found such as transient induction (class 4) and decrease in binding (class 5). Sko1 and Yap6 also bound many Yap4 constitutive targets of class 1, at either 0 min or 30 min. Another interesting aspect is that Msn2 preferentially binds inducible Yap4 targets. Moreover, Yap6 and Sko1 bind a significant number of salt-induced Yap4 targets that belong to class 2. It seems from the results of Ni et al. that the binding of other factors correlates with induced binding, and thus the association of different components at induced targets regulates gene expression [101]. Yap4 targets were involved in oxidoreductase activity and Yap4, together with Sko1, have targets involved in hexose transport, glucose and ethanol catabolism. Yap6 has targets in the same categories as Yap4 but it specifically targets genes encoding ribosomal proteins.
–
Yap4 is a highly phosphorylated protein. This post-translation modification is dependent on PKA and GSK3 and was shown to affect its stability but not its nuclear localization [102].
–
Finally, Yap4 and Yap6 were shown to interact with the general transcriptional repressor Tup1, suggesting that they could also act as transcriptional repressors [16][93].
YAP7 AND NITROSATIVE STRESS
The function of Yap7 (YOL028c) has not been completely deciphered. It was described that Yap7 represses YHB1, encoding a flavohemoglobin which functions as a nitric oxide (NO) oxidoreductase [19]. In consequence, Yap7 deletion confers high resistance to NO. Yap7 repression of YHB1 is exerted by binding YRE-O motifs in the YHB1 promoter and by recruiting the transcriptional repressor Tup1 [19][103]. Like Yap5, Yap7 has a bipartite Hap4L-bZIP domain, which was shown to play a role in its function. However, the de-repression of YHB1 observed in a mutant of CBC is only 30% of that observed in a yap7 mutant, indicating that Yap7 repressor activity is only partially dependent on CBC [19]. Noteworthy, in laboratory S. cerevisiae strains, YAP7 is interrupted by a frame-shift and produces a truncated protein which has DNA binding properties but lacks the Tup1 interaction domain and is unable to repress transcription. The role of Yap7 was therefore revealed by studying “wild” yeast strains expressing a full-length Yap7 protein [19].
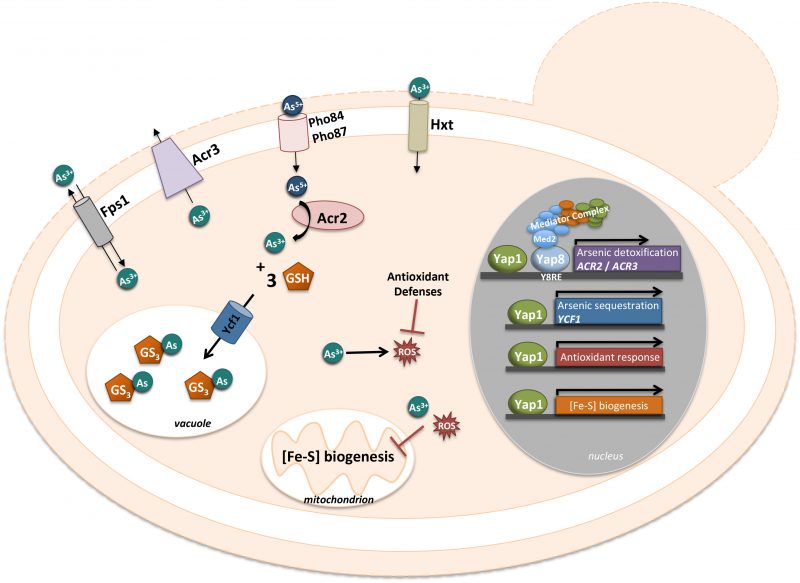
YAP8 AND ITS ROLE IN THE DETOXIFICATION OF ARSENIC COMPOUNDS
Arsenic (As) is the 20th most abundant element in the earth's crust and is a highly toxic metalloid with respect to the human health being the most potent human carcinogen. Although synonymous to a poison, it is one of the oldest drugs in the history of humankind, first used to treat syphilis and later malaria. In spite of its toxic effects, arsenic is also a chemotherapeutic agent in the treatment of the acute promyelocytic leukemia (APL) as well as other solid cancers [104][105][106][107][108]. Arsenic is a multifactorial element because this metalloid interferes with several metabolic pathways leading to a myriad of cytotoxic effects, forming ROS with the induction of apoptosis [56].
–
The global environmental widespread of arsenic led the organisms to develop and evolve several detoxification mechanisms for this compound. In S. cerevisiae the main detoxification system for arsenic is composed of Acr2/Arr2, an arsenate reductase responsible for the reduction of arsenate (As(V)) to arsenite (As(III)) [109] and Acr3/Arr3, a dual As(III) and antimonite (Sb(III)) plasma-membrane efflux protein [45][110]. The transcription factor Yap8/Arr1 is the master regulator of arsenic detoxification that binds the promoter region located between ACR2 and ACR3, two divergently transcribed genes (Fig. 4). Remarkably, the YAP8 gene itself is located just next to ACR2 and ACR3, hence forming a genomic cluster specialized in arsenic resistance. Yap8 specifically recognizes and binds an extended YRE with a 13 bp pseudo-palindromic sequence (TGATTAATAATCA), where both the core element (TTAATAA) and the flanking sequences are essential for Yap8 binding and transcriptional activation of its targets (Fig. 4) [20][45]. Yap8 was found to be regulated by arsenic at the level of its nucleo-cytoplasmic shuttling. When exposed to arsenic the cysteine residues, Cys132, Cys137 and Cys274, bind the arsenic compound masking the NES and as such, Yap8 will remain in the nucleus activating its targets [57]. However, contrary to these observations, another study showed that Yap8 is constitutively nuclear, being associated with the ACR3 promoter in untreated as well as As(III)-exposed cells [45]. Unknown genetic differences in S. cerevisiae strains used and/or different expression systems may account for the discrepancies between both works. The three conserved cysteine residues, Cys132, Cys137 and Cys274, are important for Yap8 transactivation function, since the mutants obtained by substitution for each residue failed to induce ACR3 expression [57]. Menezes et al. have shown that arsenate alters the sulfhydryl state of Yap8 conserved cysteines, suggesting that these residues may also be direct sensors of the pentavalent form of arsenic, As (V) [111].
–
–
As(V) also induced the expression of the Aft1-dependent gene CTH2 to levels similar to those triggered by BPS (bactophenantroline disulfonic acid) [112]. However, in the presence of As, the expression of the high-affinity iron uptake protein encoding genes FET3 and FTR1 is abrogated and the aft1 mutant growth is impaired. Furthermore, growth of the aft1 mutant in presence of As is totally resumed when iron is added, indicating a physiological link between As and the Aft1/2 regulon iron deprivation response [112]. As such, these data revealed that As(V) causes Fe scarcity.
–
Tamas´s laboratory showed that Yap8 escapes degradation under arsenic conditions [113]. Later, Ferreira et al. have shown how Yap8 circumvents proteolysis under As stress [114]. Although Ufd2, an E4-Ubiquitin ligase, was involved in protein degradation, those authors found that the UFD2 deletion causes Yap8 degradation and a decrease in its transcriptional activity. Consequently, the cell growth under arsenic stress is compromised in this mutant. These data suggested that Ufd2 possesses another function besides proteolysis. Several reports indicated that the Ufd2 U-box motif is essential for ubiquitination and as such, is required for Ufd2 action during proteolysis [115][116]. However, the U box motif is not functional in the Ufd2 that is acting as a stabilizing protein in the activity of Yap8 [114].
–
Another important aspect of Yap8 activity relies on its interaction with the core transcriptional machinery and more particularly with the Mediator complex (Fig. 4). The Mediator is a complex molecular machine composed of about 20 subunits organized in four domains (tail, middle, head and regulatory modules) [117][118][119]. The tail module contains the subunits Med2, Med3, Med15 and Med16 interacting with transactivators and as such recruiting the complex to the gene promoter. Until now a small numbers of associations between the Mediator tail subunits and transcription factors such as Pdr1/Pdr3 [120], Hsf1 [121], Gal4 [122], and Gcn4 [123] were described. Using two hybrid assays in the presence of arsenate, Menezes et al. revealed that Yap8 is a partner of Med2, a result afterwards confirmed by chromatin immunoprecipitation assays [111]. After Yap8 activation, the Mediator binds the ACR2/ACR3 promoter through the interaction with the mediator complex via the tail subunit, Med2 (Fig. 4). Transcription is also under the control of the SWI/SNF and SAGA chromatin-remodeling complexes [124][125][126]. In the case of Yap8, the specific SWI/SNF and SAGA subunits (Snf2, Snf5 and Spt20) are as well required for the full expression of ACR2. In conclusion, Menezes et al. showed that Yap8 is a direct sensor of arsenate and that the Mediator and chromatin-remodelers SWI/SNF and SAGA are essential coactivators for the expression of Yap8 targets, ACR2 and ACR3 [111].
THE YAP FAMILIY IN OTHER FUNGAL SPECIAS: AN EVOLUTIONARY PERSPECTIVE
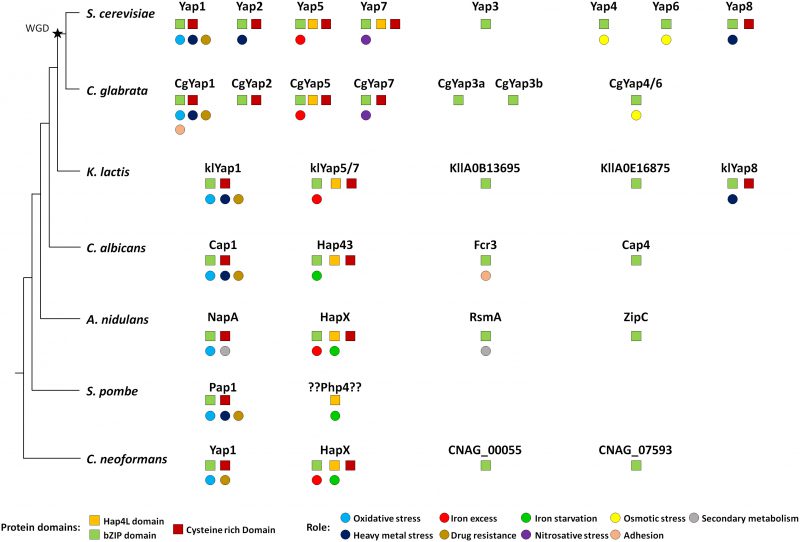
Orthologues of Yap transcription factors are found in all fungi. Most species have three to four members of this family, with the notable exception of the fission yeast Schizosaccharomyces pombe, which has only one (Fig. 5). The eight YAP genes described in S. cerevisiae actually originated from whole genome duplication (WGD), which occurred in its ancestry. This WGD created pairs of ohnologues (i.e. paralogues arising from the WGD). It was followed by massive gene reduction and most of the ohnologues were lost. Consequently, the modern yeast species which have encountered the WGD (named post-WGD species) have roughly the same number of genes than those which haven't (called pre-WGD species) [127]. Yet, in some cases, the ohnologues evolved divergent functions which were positively selected and retained. This was the case for the Yap family and most extent post-WGD yeast species have six to eight YAP genes, with some variations in the repertoire. In S. cerevisiae, three pairs of ohnologues were retained: Yap1/Yap2, Yap4/Yap6 and Yap5/Yap7 (Fig. 1). Another well-studied post-WGD species, the human pathogen Candida glabrata, has seven YAP genes: it lacks an orthologue for YAP8, has two YAP3 genes (named CgYAP3a and CgYAP3b) and only one orthologue for the YAP4 and YAP6 pair (named CgYAP4/6) (Fig. 5) [103].
–
–
Yap1 is the most conserved member of the family, both in terms of sequence and in terms of function (Fig. 5). Orthologues playing a major role in oxidative stress response have been described in post-WGD yeast species (e.g. CgYap1 in C. glabrata), pre-WGD yeast species (e.g. KlYap1 in Kluyveromyces lactis, Cap1 in the human pathogen Candida albicans), euascomycetes (e.g. NapA in Aspergillus nidulans), archeascomycetes (e.g. Pap1 in S. pombe) and basidyomycetes (e.g. Yap1 in the human pathogen Cryptococcus neoformans) [128][129][130][131][132][133][134]. Although their list of target genes diverged, they all share a core set of regulated genes which are essential for redox homeostasis (e.g. catalase, thioredoxin, thioredoxin dependent peroxydases, glutathione reductase, etc.) [12][17][135][136][137][138]. All the Yap1 orthologues recognize YRE-O motifs [12][139], but CgYap1 and Pap1 are also able to interact with YRE-A sites [17][140]. The mechanism for Yap1 redox sensing is remarkably conserved. In C. glabrata, the overexpression of Ybp1 confers resistance to oxidative stress and CgYap1 cooperates with CgSkn7 to regulate a set of protective genes under oxidative stress, as described in S. cerevisiae [136][141]. In C. albicans, Cap1 is regulated at the level of its nucleocytoplasmic localization due to the interaction of its CRDs with Ybp1 and Hyr1/Orp1/Gpx3, as described for Yap1 in S. cerevisiae [142][143]. Notably, Cap1 mutants are defective for macrophage escape and less virulent than wild-type strains [142]. In S. pombe, Crm1 actively exports Pap1 from the nucleus [134][144][145]. Pap1 is activated by the oxidation of its CRDs by the peroxiredoxin Tpx1 [146][147][148][149].
–
The involvement of Yap1 in multidrug resistance is also largely conserved (Fig. 5). Null mutants for YAP1 orthologues show sensitivity to multiple drugs in C. glabrata, K. lactis, C. albicans, S. pombe and C. neoformans [131][133][150][151][152][153]. The role of Yap1 and Yap2 in cadmium detoxification is slightly less widespread. Cadmium sensitivity has been observed for mutants of Yap1 orthologues in C. glabrata, K. lactis, C. albicans, N. crassa and S. pombe, but not in A. nidulans and C. neoformans [103][129][130][131][133][134][150][154]. Besides these conserved roles, some Yap1 orthologues have more species-specific functions. For instance, in C. glabrata, CgYap1 controls the expression of the specific adhesin Epa2, which is involved in host colonization [155]. In A. nidulans, NapA modulates secondary metabolites production and sexual development [156][157][158].
–
The Yap5 and Yap7 proteins show poor sequence conservation over the full sequence. Still, their lineage can be easily traced over large evolutionary distances due to their characteristic Hap4L-bZip bipartite domain, which provides them with the potential to interact both with the CCAAT binding complex and with DNA [73][159][160][161]. Moreover, most of them have retained the specific CRD, which is used by Yap5 to sense iron-sulfur clusters [76][162]. Orthologues have been found in C. glabrata (CgYap5 and CgYap7), K. lactis (KlYap5/7), C. albicans (Hap43) and in euascomycetes and basidyomycetes (HapX). These proteins are consistently involved in fungal iron homeostasis and their interaction with the CBC is conserved in most species and required for their function (Fig. 5) [18][19][159][160][163][164]. As for Yap1, a core set of targets involved in iron consuming is remarkably conserved between Yap5, CgYap5, Hap43 and HapX [74][103][164][165][166]. However, in contrast to the Yap1 case, their precise role has been considerably rewired during evolution. In euascomycetes, HapX is involved in both iron starvation and iron excess responses, by repressing or activating the expression of the iron consuming genes depending on the iron supply [162]. In basidyomycetes, HapX is also able to act both as a repressor and as an activator [164]. In the pre-WGD yeast C. albicans, Hap43 is mostly involved in the repression of iron consuming genes when iron is limiting and it has almost no role in the iron excess response [160][163][167]. Conversely, in C. glabrata and S. cerevisiae Yap5 is majorly involved in the iron excess response and has few impacts on the iron starvation response [18][19][73][74][103]. This role of Yap5 in the protection against iron overload is conserved in the pre-WGD yeast species K. lactis and Lachancea kluyverii [19].
–
No Hap4L-bZIP bipartite protein is found in S. pombe. However, a functional homologue of HapX, named Php4, has been described in this species. Php4 has a Hap4L domain but no bZIP. It represses the expression of iron consuming genes under iron limited conditions through its interaction with CBC, but it has no role in iron excess response and does not bind DNA directly [168][169]. In contrast to HapX and Yap5, Php4 senses iron indirectly through interaction with glutaredoxins [170]. Then, it is difficult to say if Php4 is actually a HapX orthologue, which has lost its bZip domain, or if the similar role of Php4 and HapX in iron starvation response is just an evolutionary convergence between proteins of different origins.
–
The role of Yap7, the ohnologue of Yap5 in the constitutive repression of YHB1 is conserved in post-WGD species but not in the pre-WGD species K. lactis and L. kluyverii [19]. This led to propose that this role appeared after the WGD. However, this hypothesis is challenged by the fact that YHB1 is a target of Hap43 in C. albicans, which represses its expression in a Tup1-dependent way when iron is limiting [163]. Notably, YHB1 is a heme-containing protein and therefore the activity of Yap7 is also connected to iron homeostasis. Consequently, the deletion of YAP7 confers resistance to iron overload in C. glabrata, probably due to the high expression of Yhb1 which traps iron into a protein-bound, non-toxic, form [103]. Importantly, HapX proteins are required for the pathogenesis of several fungal pathogens of human and plants [163][166][171][172], but they are dispensable for virulence in C. neoformans and in the dermatophyte Arthroderma benhamiae [164][173].
–
The mode of action and DNA interaction properties of this sub-group of Yap proteins has also diverged. In euascomicetes and in C. albicans, the interaction with CBC is predominant, the most highly enriched motif in the promoter of the targets of HapX, and Hap43 is the CCAAT motif [162][165]. Still their bZIP domain is important for their regulatory properties, but it only synergically contributes to DNA binding with a loose specificity [160][174]. In C. glabrata and S. cerevisiae, the YRE-O is the most enriched motif in the target promoters of Yap5 and it is necessary for the binding to occur [18][77][103]. In C. glabrata, the interaction with the CBC is also necessary for the activity of Yap5 on its targets and a CCAAT motif is always found close to the YRE in the promoter of its targets [73]. This aspect of Yap5 regulation has not been investigated in S. cerevisiae yet. In post-WGD species, the Yap7 lineage is apparently on the way of losing the CBC interaction. Indeed, the CBC only partly contributes to the repression properties of Yap7 in S. cerevisiae [19]. In C. glabrata, CgYap7 has even totally lost its Hap4L domain and does not require CBC for its function [19].
–
In terms of sequence, Yap3 is the second most conserved Yap after Yap1 and YAP3 orthologues can be found with good confidence from S. cerevisiae to euascomycetes (Fig. 5). Yet, in most species, no clear role could be assigned to these regulators. In C. glabrata, large-scale analyses failed to identify a biologically meaningful set of targets for Yap3a and Yap3b [103]. In C. albicans, FCR3 was initially described as a partial multicopy suppressor for the fluconazole sensitivity of a pdr1Δpdr3Δ S. cerevisiae strain. However, Fcr3 has no role in drug resistance in C. albicans. The only phenotype described for FCR3 null mutants is a decrease in adherence properties [175]. In A. nidulans and in the human pathogen Aspergillus fumigatus, RsmA stimulates secondary metabolite production and controls sexual development [158][176][177][178]. However, these processes are specific for filamentous fungi and cannot be transposed to yeasts.
–
The Yap4 and Yap6 lineage shows high sequence divergence. Still probable orthologues can be identified in almost all fungal species (Fig. 5). It is not clear if the role described for these two factors in the osmotic stress response of S. cerevisiae is conserved in other species. In C. glabrata, about 40 targets of CgYap4/6 have been identified, with no obvious connection with osmotic homeostasis. Yet, CgYAP4/6 null mutant exhibits a moderate growth defect in high salt concentration conditions [103]. The orthologues of Yap4 and Yap6 in C. albicans (Cap4) and A. nidulans (ZipC) have no functional annotation, but their potential involvement in the osmotic shock response has not been investigated to our knowledge.
–
Yap8 shows a strange and hectic conservation pattern. Orthologues are found in S. cerevisiae and two closely Saccharomyces sensu stricto species, but in no other post-WGD yeasts. Additionally, YAP8 orthologues are present in a handful of pre-WGD species, namely K. lactis, two Lachancea species (out of twelve which genomes is fully sequenced) and Torulaspora microellipsoides (information taken from www.saccharomycessensustricto.org and from gryc.inra.fr). Each time YAP8 is present in a genome, an ACR3 orthologue is found just next to it on the same chromosome, hence constituting a small genomic cluster involved in arsenic resistance. Intriguingly, although the presence of YAP8 is poorly conserved in yeasts, the sequence identity between the Yap8 orthologues is high. For instance, the S. cerevisiae and L. fermentati Yap8 proteins share 46% identity over the full sequence, despite of the large evolutionary distance separating these two species. For comparison, the S. cerevisiae and L. fermentati Yap1 proteins share only 33% identity. Is it a sign of an introgression of the YAP8 locus from one species to the other? Or has Yap8 been conserved in this particular species because of special environmental selective pressures? The current knowledge does not allow answering these questions. Importantly, the role of Yap8 in arsenite resistance and its property of direct sensing of As(III) molecules are conserved in K. lactis [179][180].
CONCLUSIONS
Data obtained in the last decade have shown that gene expression regulation under stress conditions does not involve a single transcription factor but cooperation between several such factors. For instance, RPN4, which encodes a transcriptional activator of proteasome genes, contains in its promoter multiple regulatory elements bound by Hsf1, Pdr1/Pdr3 and Yap1 [181] and Yap4/Cin5 contains also in its promoter HSE/Hsf1, STRE/Msn2/Msn4 and YRE/Yap1 elements [90]. Then, responses to stress are not linear sequences of events but rather an orchestrated phenomenon that puts at play several interconnected pathways and response elements, acting via condition and gene-specific cross-talk events. This would lead to a precise response and adaptation to the new environment. In this review, we have thus focused on the major transcription factors of the Yap family that are involved in yeast stress response. It will be important to understand how the activity of these factors is coordinated, as well as to identify the signals triggering this coordination and to determine their integration with metabolic pathways. The work by Snyder's group [101] shows the interaction of factors such as Yap4/Cin5, Yap6, Sko1, Msn2 and Msn4. Furthermore, it would be attractive to study how these transcription factors interact with each other. It could be directly or indirectly via the transcriptional machinery as it was already determined for Yap8 [111]. Another important point is that several Yap transcription factors (Yap4/Cin5, Yap5, Yap6, HapX) can act as both inducers and repressors. The precise mechanisms behind this versatile activity might constitute another line of research once all the targets of these transcription factors have been known.
REFERENCES
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D and Brown PO (2000). Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11(12): 4241-57. doi: 10.1091/mbc.11.12.4241
- Reinke AW, Baek J, Ashenberg O and Keating AE (2013). Networks of bZIP protein-protein interactions diversified over a billion years of evolution. Science 340(6133): 730-4. doi: 10.1126/science.1233465
- Jindrich K and Degnan BM (2016). The diversification of the basic leucine zipper family in eukaryotes correlates with the evolution of multicellularity. BMC Evol Biol 16: 28. doi: 10.1186/s12862-016-0598-z
- Ellenberger TE, Brandl CJ, Struhl K and Harrison SC (1992). The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: crystal structure of the protein-DNA complex. Cell 71(7): 1223-37. doi: 10.1016/s0092-8674(05)80070-4
- Glover JN and Harrison SC (1995). Crystal structure of the heterodimeric bZIP transcription factor c-Fos-c-Jun bound to DNA. Nature 373: 257-61. doi: 10.1038/373257a0
- Konig P and Richmond TJ (1993). The X-ray structure of the GCN4- bZIP bound to ATF/CREB site DNA shows the complex depends on DNA flexibility. J Mol Biol 233(1): 139-54. doi: 10.1006/jmbi.1993.1490
- Rodrigues-Pousada C, Menezes RA and Pimentel C (2010). The Yap family and its role in stress response. Yeast 27(5): 245-58. doi: 10.1002/yea.1752
- Moye-Rowley WS, Harshman KD and Parker CS (1989). Yeast YAP1 encodes a novel form of the jun family of transcriptional activator proteins. Genes Dev 3(3): 283-92. doi: 10.1101/gad.3.3.283
- Schnell N, Krems B and Entian KD (1992). The PAR1 (YAP1/SNQ3) gene of Saccharomyces cerevisiae, a c-jun homologue, is involved in oxygen metabolism. Curr Genet 21(4-5): 269-73. doi: 10.1007/bf00351681
- Bossier P, Fernandes L, Rocha D and Rodrigues-Pousada C (1993). Overexpression of YAP2, coding for a new yAP protein, and YAP1 in Saccharomyces cerevisiae alleviates growth inhibition caused by 1,10- phenanthroline. J Biol Chem 268(31): 23640-5. PMID: 8226890
- Fernandes L, Rodrigues-Pousada C and Struhl K (1997). Yap, a novel family of eight bZIP proteins in Saccharomyces cerevisiae with distinct biological functions. Mol Cell Biol 17(12): 6982-93. doi: 10.1128/mcb.17.12.6982
- Goudot C, Etchebest C, Devaux F and Lelandais G (2011). The reconstruction of condition-specific transcriptional modules provides new insights in the evolution of yeast AP-1 proteins. PLoS One 6: e20924. doi: 10.1371/journal.pone.0020924
- He XJ, Mulford KE and Fassler JS (2009). Oxidative stress function of the Saccharomyces cerevisiae Skn7 receiver domain. Eukaryot Cell 8(5): 768-78. doi: 10.1128/EC.00021-09
- Nguyen DT, Alarco AM and Raymond M (2001). Multiple Yap1p- binding sites mediate induction of the yeast major facilitator FLR1 gene in response to drugs, oxidants, and alkylating agents. J Biol Chem 276(2): 1138-45. doi: 10.1074/jbc.M008377200
- Lucau-Danila A, Lelandais G, Kozovska Z, Tanty V, Delaveau T, Devaux F and Jacq C (2005). Early expression of yeast genes affected by chemical stress. Mol Cell Biol 25(5): 1860-8. doi: 10.1128/MCB.25.5.1860-1868.2005
- Tan K, Feizi H, Luo C, Fan SH, Ravasi T and Ideker TG (2008). A systems approach to delineate functions of paralogous transcription factors: role of the Yap family in the DNA damage response. Proc Natl Acad Sci U S A 105(8): 2934-9. doi: 10.1073/pnas.0708670105
- Kuo D, Licon K, Bandyopadhyay S, Chuang R, Luo C, Catalana J, Ravasi T, Tan K and Ideker T (2010). Coevolution within a transcriptional network by compensatory trans and cis mutations. Genome Res 20(12): 1672-8. doi: 10.1101/gr.111765.110
- Li L, Bagley D, Ward DM and Kaplan J (2008). Yap5 is an iron- responsive transcriptional activator that regulates vacuolar iron storage in yeast. Mol Cell Biol 28(4): 1326-37. doi: 10.1128/MCB.01219-07
- Merhej J, Delaveau T, Guitard J, Palancade B, Hennequin C, Garcia M, Lelandais G and Devaux F (2015). Yap7 is a transcriptional repressor of nitric oxide oxidase in yeasts, which arose from neofunctionalization after whole genome duplication. Mol Microbiol 96(5): 951-72. doi: 10.1111/mmi.12983
- Amaral C, Pimentel C, Matos RG, Arraiano CM, Matzapetakis M and Rodrigues-Pousada C (2013). Two residues in the basic region of the yeast transcription factor Yap8 are crucial for its DNA-binding specificity. PLoS One 8: e83328. doi: 10.1371/journal.pone.0083328
- Ilina Y, Sloma E, Maciaszczyk-Dziubinska E, Novotny M, Thorsen M, Wysocki R and Tamas MJ (2008). Characterization of the DNA-binding motif of the arsenic-responsive transcription factor Yap8p. Biochem J 415(3): 467-75. doi: 10.1042/BJ20080713
- Vilela C, Linz B, Rodrigues-Pousada C and McCarthy JE (1998). The yeast transcription factor genes YAP1 and YAP2 are subject to differential control at the levels of both translation and mRNA stability. Nucleic Acids Res 26(5): 1150-9. doi: 10.1093/nar/26.5.1150
- Vilela C, Ramirez CV, Linz B, Rodrigues-Pousada C and McCarthy JE (1999). Post-termination ribosome interactions with the 5’UTR modulate yeast mRNA stability. EMBO J 18(11): 3139-52. doi: 10.1093/emboj/18.11.3139
- Kuge S and Jones N (1994). YAP1 dependent activation of TRX2 is essential for the response of Saccharomyces cerevisiae to oxidative stress by hydroperoxides. EMBO J 13(3): 655-64. doi: 10.1002/j.1460- 2075.1994.tb06304.x
- Wemmie JA, Wu AL, Harshman KD, Parker CS and Moye-Rowley WS (1994). Transcriptional activation mediated by the yeast AP-1 protein is required for normal cadmium tolerance. J Biol Chem 269(20): 14690-7. PMID: 8182076
- Sugiyama K, Izawa S and Inoue Y (2000). The Yap1p-dependent induction of glutathione synthesis in heat shock response of Saccharomyces cerevisiae. J Biol Chem 275(20): 15535-40. doi: 10.1074/jbc.275.20.15535
- Isoyama T, Murayama A, Nomoto A and Kuge S (2001). Nuclear import of the yeast AP-1-like transcription factor Yap1p is mediated by transport receptor Pse1p, and this import step is not affected by oxidative stress. J Biol Chem 276(24): 21863-9. doi: 10.1074/jbc.M009258200
- Yan C, Lee LH and Davis LI (1998). Crm1p mediates regulated nuclear export of a yeast AP-1-like transcription factor. EMBO J 17(24): 7416-29. doi: 10.1093/emboj/17.24.7416
- Wood MJ, Andrade EC and Storz G (2003). The redox domain of the Yap1p transcription factor contains two disulfide bonds. Biochemistry 42(41): 11982-91. doi: 10.1021/bi035003d
- Toledano MB, Delaunay A, Monceau L and Tacnet F (2004). Microbial H2O2 sensors as archetypical redox signaling modules. Trends Biochem Sci 29(7): 351-7. doi: 10.1016/j.tibs.2004.05.005
- Delaunay A, Pflieger D, Barrault MB, Vinh J and Toledano MB (2002). A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell 111(4): 471-81. doi: 10.1016/s0092- 8674(02)01048-6
- Veal EA, Ross SJ, Malakasi P, Peacock E and Morgan BA (2003). Ybp1 is required for the hydrogen peroxide-induced oxidation of the Yap1 transcription factor. J Biol Chem 278(33): 30896-904. doi: 10.1074/jbc.M303542200
- Bersweiler A, D’Autreaux B, Mazon H, Kriznik A, Belli G, Delaunay- Moisan A, Toledano MB and Rahuel-Clermont S (2017). A scaffold protein that chaperones a cysteine-sulfenic acid in H2O2 signaling. Nat Chem Biol 13(8): 909-915. doi: 10.1038/nchembio.2412
- Sikes HD (2017). Redox regulation: Scaffolding H2O2 signaling. Nat Chem Biol 13(8): 818-819. doi: 10.1038/nchembio.2432
- Azevedo D, Tacnet F, Delaunay A, Rodrigues-Pousada C and Toledano MB (2003). Two redox centers within Yap1 for H2O2 and thiol-reactive chemicals signaling. Free Radic Biol Med 35(8): 889-900. doi: 10.1016/s0891-5849(03)00434-9
- Carmel-Harel O, Stearman R, Gasch AP, Botstein D, Brown PO and Storz G (2001). Role of thioredoxin reductase in the Yap1p-dependent response to oxidative stress in Saccharomyces cerevisiae. Mol Microbiol 39(3): 595-605. doi: 10.1046/j.1365-2958.2001.02255.x
- Delaunay A, Isnard AD and Toledano MB (2000). H2O2 sensing through oxidation of the Yap1 transcription factor. EMBO J 19(19): 5157-66. doi: 10.1093/emboj/19.19.5157
- Izawa S, Maeda K, Sugiyama K, Mano J, Inoue Y and Kimura A (1999). Thioredoxin deficiency causes the constitutive activation of Yap1, an AP-1-like transcription factor in Saccharomyces cerevisiae. J Biol Chem 274(40): 28459-65. doi: 10.1074/jbc.274.40.28459
- Huang ME, Facca C, Fatmi Z, Baille D, Benakli S and Vernis L (2016). DNA replication inhibitor hydroxyurea alters Fe-S centers by producing reactive oxygen species in vivo. Sci Rep 6: 29361. doi: 10.1038/srep29361
- Vernis L, Facca C, Delagoutte E, Soler N, Chanet R, Guiard B, Faye G and Baldacci G (2009). A newly identified essential complex, Dre2- Tah18, controls mitochondria integrity and cell death after oxidative stress in yeast. PLoS One 4: e4376. doi: 10.1371/journal.pone.0004376
- Mandon EC, Trueman SF and Gilmore R (2013). Protein translocation across the rough endoplasmic reticulum. Cold Spring Harb Perspect Biol 5(2): a013342. doi: 10.1101/cshperspect.a013342
- Ponsero AJ, Igbaria A, Darch MA, Miled S, Outten CE, Winther JR, Palais G, D’Autreaux B, Delaunay-Moisan A and Toledano MB (2017). Endoplasmic Reticulum Transport of Glutathione by Sec61 Is Regulated by Ero1 and Bip. Mol Cell 67(6): 962-973 e5. doi: 10.1016/j.molcel.2017.08.012
- Jomova K and Valko M (2011). Advances in metal-induced oxidative stress and human disease. Toxicology 283(2-3): 65-87. doi: 10.1016/j.tox.2011.03.001
- Summers AO (2009). Damage control: regulating defenses against toxic metals and metalloids. Curr Opin Microbiol 12(2): 138-44. doi: 10.1016/j.mib.2009.02.003
- Wysocki R and Tamas MJ (2010). How Saccharomyces cerevisiae copes with toxic metals and metalloids. FEMS Microbiol Rev 34(6): 925-51. doi: 10.1111/j.1574-6976.2010.00217.x
- Hartwig A and Schwerdtle T (2002). Interactions by carcinogenic metal compounds with DNA repair processes: toxicological implications. Toxicol Lett 127(1-3): 47-54. doi: 10.1016/s0378- 4274(01)00482-9
- Kasprzak KS (1995). Possible role of oxidative damage in metal- induced carcinogenesis. Cancer Invest 13(4): 411-30. doi: 10.3109/07357909509031921
- Ghosh M, Shen J and Rosen BP (1999). Pathways of As(III) detoxification in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 96(9): 5001-6. doi: 10.1073/pnas.96.9.5001
- Wemmie JA, Szczypka MS, Thiele DJ and Moye-Rowley WS (1994). Cadmium tolerance mediated by the yeast AP-1 protein requires the presence of an ATP-binding cassette transporter-encoding gene, YCF1. J Biol Chem 269(51): 32592-7. PMID: 7798263
- Valko M, Morris H and Cronin MT (2005). Metals, toxicity and oxidative stress. Curr Med Chem 12(10): 1161-208. doi: 10.2174/0929867053764635
- Pimentel C, Caetano SM, Menezes R, Figueira I, Santos CN, Ferreira RB, Santos MA and Rodrigues-Pousada C (2014). Yap1 mediates tolerance to cobalt toxicity in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta 1840(6): 1977-86. doi: 10.1016/j.bbagen.2014.01.032
- Jensen LT, Ajua-Alemanji M and Culotta VC (2003). The Saccharomyces cerevisiae high affinity phosphate transporter encoded by PHO84 also functions in manganese homeostasis. J Biol Chem 278(43): 42036-40. doi: 10.1074/jbc.M307413200
- Culotta VC and Daly MJ (2013). Manganese complexes: diverse metabolic routes to oxidative stress resistance in prokaryotes and yeast. Antioxid Redox Signal 19(9): 933-44. doi: 10.1089/ars.2012.5093
- McNaughton RL, Reddi AR, Clement MH, Sharma A, Barnese K, Rosenfeld L, Gralla EB, Valentine JS, Culotta VC and Hoffman BM (2010). Probing in vivo Mn2+ speciation and oxidative stress resistance in yeast cells with electron-nuclear double resonance spectroscopy. Proc Natl Acad Sci U S A 107(35): 15335-9. doi: 10.1073/pnas.1009648107
- Caetano SM, Menezes R, Amaral C, Rodrigues-Pousada C and Pimentel C (2015). Repression of the Low Affinity Iron Transporter Gene FET4: A NOVEL MECHANISM AGAINST CADMIUM TOXICITY ORCHESTRATED BY YAP1 VIA ROX1. J Biol Chem 290(30): 18584-95. doi: 10.1074/jbc.M114.600742
- Menezes RA, Amaral C, Batista-Nascimento L, Santos C, Ferreira RB, Devaux F, Eleutherio EC and Rodrigues-Pousada C (2008). Contribution of Yap1 towards Saccharomyces cerevisiae adaptation to arsenic-mediated oxidative stress. Biochem J 414(2): 301-11. doi: 10.1042/BJ20071537
- Menezes RA, Amaral C, Delaunay A, Toledano M and Rodrigues- Pousada C (2004). Yap8p activation in Saccharomyces cerevisiae under arsenic conditions. FEBS Lett 566(1-3): 141-6. doi: 10.1016/j.febslet.2004.04.019
- Wysocki R, Fortier PK, Maciaszczyk E, Thorsen M, Leduc A, Odhagen A, Owsianik G, Ulaszewski S, Ramotar D and Tamas MJ (2004). Transcriptional activation of metalloid tolerance genes in Saccharomyces cerevisiae requires the AP-1-like proteins Yap1p and Yap8p. Mol Biol Cell 15(5): 2049-60. doi: 10.1091/mbc.e03-04-0236
- da Silva SM, Batista-Nascimento L, Gaspar-Cordeiro A, Vernis L, Pimentel C and Rodrigues-Pousada C (2018). Transcriptional regulation of FeS biogenesis genes: A possible shield against arsenate toxicity activated by Yap1. Biochim Biophys Acta Gen Subj 1862(10): 2152-2161. doi: 10.1016/j.bbagen.2018.07.013
- Stephen DW, Rivers SL and Jamieson DJ (1995). The role of the YAP1 and YAP2 genes in the regulation of the adaptive oxidative stress responses of Saccharomyces cerevisiae. Mol Microbiol 16(3): 415-23. doi: 10.1111/j.1365-2958.1995.tb02407.x
- Cohen BA, Pilpel Y, Mitra RD and Church GM (2002). Discrimination between paralogs using microarray analysis: application to the Yap1p and Yap2p transcriptional networks. Mol Biol Cell 13(5): 1608-14. doi: 10.1091/mbc.01-10-0472
- Azevedo D, Nascimento L, Labarre J, Toledano MB and Rodrigues- Pousada C (2007). The S. cerevisiae Yap1 and Yap2 transcription factors share a common cadmium-sensing domain. FEBS Lett 581(12): 187-95. doi: 10.1016/j.febslet.2006.11.083
- Hirata D, Yano K and Miyakawa T (1994). Stress-induced transcriptional activation mediated by YAP1 and YAP2 genes that encode the Jun family of transcriptional activators in Saccharomyces cerevisiae. Mol Gen Genet 242(3): 250-6. doi: 10.1007/bf00280413
- McHale MW, Kroening KD and Bernlohr DA (1996). Identification of a class of Saccharomyces cerevisiae mutants defective in fatty acid repression of gene transcription and analysis of the frm2 gene. Yeast 12(4): 319-31. doi: 10.1002/(SICI)1097-0061(19960330)12:4%3C319::AID-YEA914%3E3.0.CO;2-#
- Ball CA, Dolinski K, Dwight SS, Harris MA, Issel-Tarver L, Kasarskis A, Scafe CR, Sherlock G, Binkley G, Jin H, Kaloper M, Orr SD, Schroeder M, Weng S, Zhu Y, Botstein D and Cherry JM (2000). Integrating functional genomic information into the Saccharomyces genome database. Nucleic Acids Res 28(1): 77-80. doi: 10.1093/nar/28.1.77
- Howlett NG and Avery SV (1997). Induction of lipid peroxidation during heavy metal stress in Saccharomyces cerevisiae and influence of plasma membrane fatty acid unsaturation. Appl Environ Microbiol 63(8): 2971-6. PMID: 9251184
- Bilsland E, Molin C, Swaminathan S, Ramne A and Sunnerhagen P (2004). Rck1 and Rck2 MAPKAP kinases and the HOG pathway are required for oxidative stress resistance. Mol Microbiol 53(6): 1743-56. doi: 10.1111/j.1365-2958.2004.04238.x
- Chang M, Kang HJ, Baek IJ, Kang CM, Park YS and Yun CW (2013). Rck1 up-regulates Hog1 activity by down-regulating Slt2 activity in Saccharomyces cerevisiae. Biochem Biophys Res Commun 440(1): 119-24. doi: 10.1016/j.bbrc.2013.09.045
- Mazzola D, Pimentel C, Caetano S, Amaral C, Menezes R, Santos CN, Eleutherio E and Rodrigues-Pousada C (2015). Inhibition of Yap2 activity by MAPKAP kinase Rck1 affects yeast tolerance to cadmium. FEBS Lett 589(19 Pt B): 2841-9. doi: 10.1016/j.febslet.2015.07.049
- North M, Tandon VJ, Thomas R, Loguinov A, Gerlovina I, Hubbard AE, Zhang L, Smith MT and Vulpe CD (2011). Genome-wide functional profiling reveals genes required for tolerance to benzene metabolites in yeast. PLoS One 6: e24205. doi: 10.1371/journal.pone.0024205
- Tan SX, Teo M, Lam YT, Dawes IW and Perrone GG (2009). Cu, Zn superoxide dismutase and NADP(H) homeostasis are required for tolerance of endoplasmic reticulum stress in Saccharomyces cerevisiae. Mol Biol Cell 20(5): 1493-508. doi: 10.1091/mbc.E08-07- 0697
- Jo WJ, Loguinov A, Wintz H, Chang M, Smith AH, Kalman D, Zhang L, Smith MT and Vulpe CD (2009). Comparative functional genomic analysis identifies distinct and overlapping sets of genes required for resistance to monomethylarsonous acid (MMAIII) and arsenite (AsIII) in yeast. Toxicol Sci 111(2): 424-36. doi: 10.1093/toxsci/kfp162
- Thiebaut A, Delaveau T, Benchouaia M, Boeri J, Garcia M, Lelandais G and Devaux F (2017). The CCAAT-Binding Complex Controls Respiratory Gene Expression and Iron Homeostasis in Candida Glabrata. Sci Rep 7: 3531. doi: 10.1038/s41598-017-03750-5
- Pimentel C, Vicente C, Menezes RA, Caetano S, Carreto L and Rodrigues-Pousada C (2012). The role of the Yap5 transcription factor in remodeling gene expression in response to Fe bioavailability. PLoS One 7: e37434. doi: 10.1371/journal.pone.0037434
- Li L, Kaplan J and Ward DM (2017). The glucose sensor Snf1 and the transcription factors Msn2 and Msn4 regulate transcription of the vacuolar iron importer gene CCC1 and iron resistance in yeast. J Biol Chem 292(37): 15577-15586. doi: 10.1074/jbc.M117.802504
- Rietzschel N, Pierik AJ, Bill E, Lill R and Muhlenhoff U (2015). The basic leucine zipper stress response regulator Yap5 senses high-iron conditions by coordination of [2Fe-2S] clusters. Mol Cell Biol 35(2): 370-8. doi: 10.1128/MCB.01033-14
- Li L, Jia X, Ward DM and Kaplan J (2011). Yap5 protein-regulated transcription of the TYW1 gene protects yeast from high iron toxicity. J Biol Chem 286(44): 38488-97. doi: 10.1074/jbc.M111.286666
- Lin H, Li L, Jia X, Ward DM and Kaplan J (2011). Genetic and biochemical analysis of high iron toxicity in yeast: iron toxicity is due to the accumulation of cytosolic iron and occurs under both aerobic and anaerobic conditions. J Biol Chem 286(5): 3851-62. doi: 10.1074/jbc.M110.190959
- Noma A, Kirino Y, Ikeuchi Y and Suzuki T (2006). Biosynthesis of wybutosine, a hyper-modified nucleoside in eukaryotic phenylalanine tRNA. EMBO J 25(10): 2142-54. doi: 10.1038/sj.emboj.7601105
- Ojeda L, Keller G, Muhlenhoff U, Rutherford JC, Lill R and Winge DR (2006). Role of glutaredoxin-3 and glutaredoxin-4 in the iron regulation of the Aft1 transcriptional activator in Saccharomyces cerevisiae. J Biol Chem 281(26): 17661-9. doi: 10.1074/jbc.M602165200
- Pujol-Carrion N, Belli G, Herrero E, Nogues A and de la Torre-Ruiz MA (2006). Glutaredoxins Grx3 and Grx4 regulate nuclear localisation of Aft1 and the oxidative stress response in Saccharomyces cerevisiae. J Cell Sci 119(Pt 21): 4554-64. doi: 10.1242/jcs.03229
- Li L, Miao R, Bertram S, Jia X, Ward DM and Kaplan J (2012). A role for iron-sulfur clusters in the regulation of transcription factor Yap5- dependent high iron transcriptional responses in yeast. J Biol Chem 287(42): 35709-21. doi: 10.1074/jbc.M112.395533
- Saito H and Posas F (2012). Response to hyperosmotic stress. Genetics 192(2): 289-318. doi: 10.1534/genetics.112.140863
- Reed RH, Chudek JA, Foster R and Gadd GM (1987). Osmotic significance of glycerol accumulation in exponentially growing yeasts. Appl Environ Microbiol 53(9): 2119-23. PMID: 3314706
- Yancey PH, Clark ME, Hand SC, Bowlus RD and Somero GN (1982). Living with water stress: evolution of osmolyte systems. Science 217(4566): 1214-22. doi: 10.1126/science.7112124
- Proft M, Pascual-Ahuir A, de Nadal E, Arino J, Serrano R and Posas F (2001). Regulation of the Sko1 transcriptional repressor by the Hog1 MAP kinase in response to osmotic stress. EMBO J 20(5): 1123-33. doi: 10.1093/emboj/20.5.1123
- Rep M, Reiser V, Gartner U, Thevelein JM, Hohmann S, Ammerer G and Ruis H (1999). Osmotic stress-induced gene expression in Saccharomyces cerevisiae requires Msn1p and the novel nuclear factor Hot1p. Mol Cell Biol 19(8): 5474-85. doi: 10.1128/mcb.19.8.5474
- Rep M, Krantz M, Thevelein JM and Hohmann S (2000). The transcriptional response of Saccharomyces cerevisiae to osmotic shock. Hot1p and Msn2p/Msn4p are required for the induction of subsets of high osmolarity glycerol pathway-dependent genes. J Biol Chem 275(12): 8290-300. doi: 10.1074/jbc.275.12.8290
- Hoyt MA, Stearns T and Botstein D (1990). Chromosome instability mutants of Saccharomyces cerevisiae that are defective in microtubule-mediated processes. Mol Cell Biol 10(1): 223-34. doi: 10.1128/mcb.10.1.223
- Nevitt T, Pereira J, Azevedo D, Guerreiro P and Rodrigues-Pousada C (2004). Expression of YAP4 in Saccharomyces cerevisiae under osmotic stress. Biochem J 379(Pt 2): 367-74. doi: 10.1042/BJ20031127
- Delling U, Raymond M and Schurr E (1998). Identification of Saccharomyces cerevisiae genes conferring resistance to quinoline ring-containing antimalarial drugs. Antimicrob Agents Chemother 42(5): 1034-41. doi: 10.1128/aac.42.5.1034
- Furuchi T, Ishikawa H, Miura N, Ishizuka M, Kajiya K, Kuge S and Naganuma A (2001). Two nuclear proteins, Cin5 and Ydr259c, confer resistance to cisplatin in Saccharomyces cerevisiae. Mol Pharmacol 59(3): 470-4. doi: 10.1124/mol.59.3.470
- Hanlon SE, Rizzo JM, Tatomer DC, Lieb JD and Buck MJ (2011). The stress response factors Yap6, Cin5, Phd1, and Skn7 direct targeting of the conserved co-repressor Tup1-Ssn6 in S. cerevisiae. PLoS One 6: e19060. doi: 10.1371/journal.pone.0019060
- Nevitt T, Pereira J and Rodrigues-Pousada C (2004). YAP4 gene expression is induced in response to several forms of stress in Saccharomyces cerevisiae. Yeast 21(16): 1365-74. doi: 10.1002/yea.1188
- Sollner S, Nebauer R, Ehammer H, Prem A, Deller S, Palfey BA, Daum G and Macheroux P (2007). Lot6p from Saccharomyces cerevisiae is a FMN-dependent reductase with a potential role in quinone detoxification. FEBS J 274(5): 1328-39. doi: 10.1111/j.1742- 4658.2007.05682.x
- Sollner S, Deller S, Macheroux P and Palfey BA (2009). Mechanism of flavin reduction and oxidation in the redox-sensing quinone reductase Lot6p from Saccharomyces cerevisiae. Biochemistry 48(36): 8636-43. doi: 10.1021/bi900734a
- Sollner S and Macheroux P (2009). New roles of flavoproteins in molecular cell biology: an unexpected role for quinone reductases as regulators of proteasomal degradation. FEBS J 276(16): 4313-24. doi: 10.1111/j.1742-4658.2009.07143.x
- Sollner S, Schober M, Wagner A, Prem A, Lorkova L, Palfey BA, Groll M and Macheroux P (2009). Quinone reductase acts as a redox switch of the 20S yeast proteasome. EMBO Rep 10(19): 65-70. doi: 10.1038/embor.2008.218
- Borneman AR, Gianoulis TA, Zhang ZD, Yu H, Rozowsky J, Seringhaus MR, Wang LY, Gerstein M and Snyder M (2007). Divergence of transcription factor binding sites across related yeast species. Science 317(5839): 815-9. doi: 10.1126/science.1140748
- Horak CE, Luscombe NM, Qian J, Bertone P, Piccirrillo S, Gerstein M and Snyder M (2002). Complex transcriptional circuitry at the G1/S transition in Saccharomyces cerevisiae. Genes Dev 16(23): 3017-33. doi: 10.1101/gad.1039602
- Ni L, Bruce C, Hart C, Leigh-Bell J, Gelperin D, Umansky L, Gerstein MB and Snyder M (2009). Dynamic and complex transcription factor binding during an inducible response in yeast. Genes Dev 23(11): 1351-63. doi: 10.1101/gad.1781909
- Pereira J, Pimentel C, Amaral C, Menezes RA and Rodrigues- Pousada C (2009). Yap4 PKA- and GSK3-dependent phosphorylation affects its stability but not its nuclear localization. Yeast 26(12): 641- 53. doi: 10.1002/yea.1711
- Merhej J, Thiebaut A, Blugeon C, Pouch J, Ali Chaouche Mel A, Camadro JM, Le Crom S, Lelandais G and Devaux F (2016). A Network of Paralogous Stress Response Transcription Factors in the Human Pathogen Candida glabrata. Front Microbiol 7: 645. doi: 10.3389/fmicb.2016.00645
- Davison K, Cote S, Mader S and Miller WH (2003). Glutathione depletion overcomes resistance to arsenic trioxide in arsenic-resistant cell lines. Leukemia 17(5): 931-40. doi: 10.1038/sj.leu.2402876
- Hu J, Liu YF, Wu CF, Xu F, Shen ZX, Zhu YM, Li JM, Tang W, Zhao WL, Wu W, Sun HP, Chen QS, Chen B, Zhou GB, Zelent A, Waxman S, Wang ZY, Chen SJ and Chen Z (2009). Long-term efficacy and safety of all-trans retinoic acid/arsenic trioxide-based therapy in newly diagnosed acute promyelocytic leukemia. Proc Natl Acad Sci U S A 106(9): 3342-7. doi: 10.1073/pnas.0813280106
- Patrick L (2003). Toxic metals and antioxidants: Part II. The role of antioxidants in arsenic and cadmium toxicity. Altern Med Rev 8(2): 106-28. PMID: 12777158
- Rosen BP (2002). Biochemistry of arsenic detoxification. FEBS Lett 529(1): 86-92. doi: 10.1016/s0014-5793(02)03186-1
- Rosen BP (2002). Transport and detoxification systems for transition metals, heavy metals and metalloids in eukaryotic and prokaryotic microbes. Comp Biochem Physiol A Mol Integr Physiol 133(3): 689-93. doi: 10.1016/s1095-6433(02)00201-5
- Mukhopadhyay R and Rosen BP (1998). Saccharomyces cerevisiae ACR2 gene encodes an arsenate reductase. FEMS Microbiol Lett 168(1): 127-36. doi: 10.1111/j.1574-6968.1998.tb13265.x
- Maciaszczyk-Dziubinska E, Migdal I, Migocka M, Bocer T and Wysocki R (2010). The yeast aquaglyceroporin Fps1p is a bidirectional arsenite channel. FEBS Lett 584(4): 726-32. doi: 10.1016/j.febslet.2009.12.027
- Menezes RA, Pimentel C, Silva AR, Amaral C, Merhej J, Devaux F and Rodrigues-Pousada C (2017). Mediator, SWI/SNF and SAGA complexes regulate Yap8-dependent transcriptional activation of ACR2 in response to arsenate. Biochim Biophys Acta Gene Regul Mech 1860(4): 472-481. doi: 10.1016/j.bbagrm.2017.02.001
- Batista-Nascimento L, Toledano MB, Thiele DJ and Rodrigues- Pousada C (2013). Yeast protective response to arsenate involves the repression of the high affinity iron uptake system. Biochim Biophys Acta 1833(5): 997-1005. doi: 10.1016/j.bbamcr.2012.12.018
- Di Y and Tamas MJ (2007). Regulation of the arsenic-responsive transcription factor Yap8p involves the ubiquitin-proteasome pathway. J Cell Sci 120(Pt 2): 256-64. doi: 10.1242/jcs.03346
- Ferreira RT, Menezes RA and Rodrigues-Pousada C (2015). E4- Ubiquitin ligase Ufd2 stabilizes Yap8 and modulates arsenic stress responses independent of the U-box motif. Biol Open 4(9): 1122-31. doi: 10.1242/bio.010405
- Aravind L and Koonin EV (2000). The U box is a modified RING finger – a common domain in ubiquitination. Curr Biol 10(4): R132-4. doi: 10.1016/s0960-9822(00)00398-5
- Tu D, Li W, Ye Y and Brunger AT (2007). Structure and function of the yeast U-box-containing ubiquitin ligase Ufd2p. Proc Natl Acad Sci U S A 104(40): 15599-606. doi: 10.1073/pnas.0701369104
- Conaway RC and Conaway JW (2011). Origins and activity of the Mediator complex. Semin Cell Dev Biol 22(7): 729-34. doi: 10.1016/j.semcdb.2011.07.021
- Kelleher RJ, 3rd, Flanagan PM and Kornberg RD (1990). A novel mediator between activator proteins and the RNA polymerase II transcription apparatus. Cell 61(7): 1209-15. doi: 10.1016/0092- 8674(90)90685-8
- Poss ZC, Ebmeier CC and Taatjes DJ (2013). The Mediator complex and transcription regulation. Crit Rev Biochem Mol Biol 48(6): 575-608. doi: 10.3109/10409238.2013.840259
- Shahi P, Gulshan K, Naar AM and Moye-Rowley WS (2010). Differential roles of transcriptional mediator subunits in regulation of multidrug resistance gene expression in Saccharomyces cerevisiae. Mol Biol Cell 21(14): 2469-82. doi: 10.1091/mbc.E09-10-0899
- Kim S and Gross DS (2013). Mediator recruitment to heat shock genes requires dual Hsf1 activation domains and mediator tail subunits Med15 and Med16. J Biol Chem 288(17): 12197-213. doi: 10.1074/jbc.M112.449553
- Ang K, Ee G, Ang E, Koh E, Siew WL, Chan YM, Nur S, Tan YS and Lehming N (2012). Mediator acts upstream of the transcriptional activator Gal4. PLoS Biol 10(3): e1001290. doi: 10.1371/journal.pbio.1001290
- Herbig E, Warfield L, Fish L, Fishburn J, Knutson BA, Moorefield B, Pacheco D and Hahn S (2010). Mechanism of Mediator recruitment by tandem Gcn4 activation domains and three Gal11 activator-binding domains. Mol Cell Biol 30(10): 2376-90. doi: 10.1128/MCB.01046-09
- Timmers HT and Tora L (2005). SAGA unveiled. Trends Biochem Sci 30(1): 7-10. doi: 10.1016/j.tibs.2004.11.007
- Wilson CJ, Chao DM, Imbalzano AN, Schnitzler GR, Kingston RE and Young RA (1996). RNA polymerase II holoenzyme contains SWI/SNF regulators involved in chromatin remodeling. Cell 84(2): 235- 44. doi: 10.1016/s0092-8674(00)80978-2
- Yudkovsky N, Logie C, Hahn S and Peterson CL (1999). Recruitment of the SWI/SNF chromatin remodeling complex by transcriptional activators. Genes Dev 13(18): 2369-74. doi: 10.1101/gad.13.18.2369
- Wolfe KH (2015). Origin of the Yeast Whole-Genome Duplication. PLoS Biol 13(8): e1002221. doi: 10.1371/journal.pbio.1002221
- Alarco AM, Balan I, Talibi D, Mainville N and Raymond M (1997). AP1-mediated multidrug resistance in Saccharomyces cerevisiae requires FLR1 encoding a transporter of the major facilitator superfamily. J Biol Chem 272(31): 19304-13. doi: 10.1074/jbc.272.31.19304
- Asano Y, Hagiwara D, Yamashino T and Mizuno T (2007). Characterization of the bZip-type transcription factor NapA with reference to oxidative stress response in Aspergillus nidulans. Biosci Biotechnol Biochem 71(7): 1800-3. doi: 10.1271/bbb.70133
- Billard P, Dumond H and Bolotin-Fukuhara M (1997). Characterization of an AP-1-like transcription factor that mediates an oxidative stress response in Kluyveromyces lactis. Mol Gen Genet 257(1): 62-70. doi: 10.1007/s004380050624
- Chen KH, Miyazaki T, Tsai HF and Bennett JE (2007). The bZip transcription factor Cgap1p is involved in multidrug resistance and required for activation of multidrug transporter gene CgFLR1 in Candida glabrata. Gene 386(1-2): 63-72. doi: 10.1016/j.gene.2006.08.010
- Lelandais G, Tanty V, Geneix C, Etchebest C, Jacq C and Devaux F (2008). Genome adaptation to chemical stress: clues from comparative transcriptomics in Saccharomyces cerevisiae and Candida glabrata. Genome Biol 9(11): R164. doi: 10.1186/gb-2008-9-11-r164
- Paul S, Doering TL and Moye-Rowley WS (2015). Cryptococcus neoformans Yap1 is required for normal fluconazole and oxidative stress resistance. Fungal Genet Biol 74: 1-9. doi: 10.1016/j.fgb.2014.10.015
- Toone WM, Kuge S, Samuels M, Morgan BA, Toda T and Jones N (1998). Regulation of the fission yeast transcription factor Pap1 by oxidative stress: requirement for the nuclear export factor Crm1 (Exportin) and the stress-activated MAP kinase Sty1/Spc1. Genes Dev 12(10): 1453-63. doi: 10.1101/gad.12.10.1453
- Chen D, Wilkinson CR, Watt S, Penkett CJ, Toone WM, Jones N and Bahler J (2008). Multiple pathways differentially regulate global oxidative stress responses in fission yeast. Mol Biol Cell 19(1): 308-17. doi: 10.1091/mbc.e07-08-0735
- Roetzer A, Klopf E, Gratz N, Marcet-Houben M, Hiller E, Rupp S, Gabaldon T, Kovarik P and Schuller C (2011). Regulation of Candida glabrata oxidative stress resistance is adapted to host environment. FEBS Lett 585(2): 319-27. doi: 10.1016/j.febslet.2010.12.006
- Thon M, Al Abdallah Q, Hortschansky P, Scharf DH, Eisendle M, Haas H and Brakhage AA (2010). The CCAAT-binding complex coordinates the oxidative stress response in eukaryotes. Nucleic Acids Res 38(4): 1098-113. doi: 10.1093/nar/gkp1091
- Znaidi S, Barker KS, Weber S, Alarco AM, Liu TT, Boucher G, Rogers PD and Raymond M (2009). Identification of the Candida albicans Cap1p regulon. Eukaryot Cell 8(6): 806-20. doi: 10.1128/EC.00002-09
- Shimanuki M, Saka Y, Yanagida M and Toda T (1995). A novel essential fission yeast gene pad1+ positively regulates pap1(+)- dependent transcription and is implicated in the maintenance of chromosome structure. J Cell Sci 108 (Pt 2): 569-79. PMID: 7769002
- Fujii Y, Shimizu T, Toda T, Yanagida M and Hakoshima T (2000). Structural basis for the diversity of DNA recognition by bZIP transcription factors. Nat Struct Biol 7(10): 889-93. doi: 10.1038/82822
- Gulshan K, Lee SS and Moye-Rowley WS (2011). Differential oxidant tolerance determined by the key transcription factor Yap1 is controlled by levels of the Yap1-binding protein, Ybp1. J Biol Chem 286(39): 34071-81. doi: 10.1074/jbc.M111.251298
- Patterson MJ, McKenzie CG, Smith DA, da Silva Dantas A, Sherston S, Veal EA, Morgan BA, MacCallum DM, Erwig LP and Quinn J (2013). Ybp1 and Gpx3 signaling in Candida albicans govern hydrogen peroxide-induced oxidation of the Cap1 transcription factor and macrophage escape. Antioxid Redox Signal 19(18): 2244-60. doi: 10.1089/ars.2013.5199
- Zhang X, De Micheli M, Coleman ST, Sanglard D and Moye- Rowley WS (2000). Analysis of the oxidative stress regulation of the Candida albicans transcription factor, Cap1p. Mol Microbiol 36(3): 618-29. doi: 10.1046/j.1365-2958.2000.01877.x
- Kuge S, Toda T, Iizuka N and Nomoto A (1998). Crm1 (XpoI) dependent nuclear export of the budding yeast transcription factor yAP-1 is sensitive to oxidative stress. Genes Cells 3(8): 521-32. doi: 10.1046/j.1365-2443.1998.00209.x
- Toda T, Shimanuki M, Saka Y, Yamano H, Adachi Y, Shirakawa M, Kyogoku Y and Yanagida M (1992). Fission yeast pap1-dependent transcription is negatively regulated by an essential nuclear protein, crm1. Mol Cell Biol 12(12): 5474-84. doi: 10.1128/mcb.12.12.5474
- Bozonet SM, Findlay VJ, Day AM, Cameron J, Veal EA and Morgan BA (2005). Oxidation of a eukaryotic 2-Cys peroxiredoxin is a molecular switch controlling the transcriptional response to increasing levels of hydrogen peroxide. J Biol Chem 280(24): 23319-27. doi: 10.1074/jbc.M502757200
- Calvo IA, Ayte J and Hidalgo E (2013). Reversible thiol oxidation in the H2O2-dependent activation of the transcription factor Pap1. J Cell Sci 126(Pt10): 2279-84. doi: 10.1242/jcs.124370
- Calvo IA, Boronat S, Domenech A, Garcia-Santamarina S, Ayte J and Hidalgo E (2013). Dissection of a redox relay: H2O2-dependent activation of the transcription factor Pap1 through the peroxidatic Tpx1-thioredoxin cycle. Cell Rep 5(5): 1413-24. doi: 10.1016/j.celrep.2013.11.027
- Vivancos AP, Castillo EA, Biteau B, Nicot C, Ayte J, Toledano MB and Hidalgo E (2005). A cysteine-sulfinic acid in peroxiredoxin regulates H2O2-sensing by the antioxidant Pap1 pathway. Proc Natl Acad Sci U S A 102(25): 8875-80. doi: 10.1073/pnas.0503251102
- Alarco AM and Raymond M (1999). The bZip transcription factor Cap1p is involved in multidrug resistance and oxidative stress response in Candida albicans. J Bacteriol 181(3): 700-8. PMID: 9922230
- Arioka M, Kouhashi M, Yoda K, Takatsuki A, Yamasaki M and Kitamoto K (1998). Multidrug resistance phenotype conferred by overexpressing bfr2+/pad1+/sks1+ or pap1+ genes and mediated by bfr1+ gene product, a structural and functional homologue of P- glycoprotein in Schizosaccharomyces pombe. Biosci Biotechnol Biochem 62(2): 390-2. doi: 10.1271/bbb.62.390
- Imrichova D, Sarinova M, Cernicka J, Gbelska Y and Subik J (2005). YAP1-mediated KNQ1 expression in Kluyveromyces lactis. FEMS Yeast Res 5(4-5): 323-9. doi: 10.1016/j.femsyr.2004.11.004
- Toda T, Shimanuki M and Yanagida M (1991). Fission yeast genes that confer resistance to staurosporine encode an AP-1-like transcription factor and a protein kinase related to the mammalian ERK1/MAP2 and budding yeast FUS3 and KSS1 kinases. Genes Dev 5(1): 60-73. doi: 10.1101/gad.5.1.60
- Tian C, Li J and Glass NL (2011). Exploring the bZIP transcription factor regulatory network in Neurospora crassa. Microbiology 157(Pt 3): 747-59. doi: 10.1099/mic.0.045468-0
- Juarez-Cepeda J, Orta-Zavalza E, Canas-Villamar I, Arreola-Gomez J, Perez-Cornejo GP, Hernandez-Carballo CY, Gutierrez-Escobedo G, Castano I and De Las Penas A (2015). The EPA2 adhesin encoding gene is responsive to oxidative stress in the opportunistic fungal pathogen Candida glabrata. Curr Genet 61(4): 529-44. doi: 10.1007/s00294-015- 0473-2
- Mendoza-Martinez AE, Lara-Rojas F, Sanchez O and Aguirre J (2017). NapA Mediates a Redox Regulation of the Antioxidant Response, Carbon Utilization and Development in Aspergillus nidulans. Front Microbiol 8: 516. doi: 10.3389/fmicb.2017.00516
- Yin WB, Reinke AW, Szilagyi M, Emri T, Chiang YM, Keating AE, Pocsi I, Wang CC and Keller NP (2013). bZIP transcription factors affecting secondary metabolism, sexual development and stress responses in Aspergillus nidulans. Microbiology 159(Pt 1): 77-88. doi: 10.1099/mic.0.063370-0
- Zheng H, Kim J, Liew M, Yan JK, Herrera O, Bok JW, Kelleher NL, Keller NP and Wang Y (2015). Redox metabolites signal polymicrobial biofilm development via the NapA oxidative stress cascade in Aspergillus. Curr Biol 25(1): 29-37. doi: 10.1016/j.cub.2014.11.018
- Hortschansky P, Eisendle M, Al-Abdallah Q, Schmidt AD, Bergmann S, Thon M, Kniemeyer O, Abt B, Seeber B, Werner ER, Kato M, Brakhage AA and Haas H (2007). Interaction of HapX with the CCAAT-binding complex–a novel mechanism of gene regulation by iron. EMBO J 26(13): 3157-68. doi: 10.1038/sj.emboj.7601752
- Singh RP, Prasad HK, Sinha I, Agarwal N and Natarajan K (2011). Cap2-HAP complex is a critical transcriptional regulator that has dual but contrasting roles in regulation of iron homeostasis in Candida albicans. J Biol Chem 286(28): 25154-70. doi: 10.1074/jbc.M111.233569
- Tanaka A, Kato M, Nagase T, Kobayashi T and Tsukagoshi N (2002). Isolation of genes encoding novel transcription factors which interact with the Hap complex from Aspergillus species. Biochim Biophys Acta 1576(1-2): 176-82. doi: 10.1016/s0167-4781(02)00286-5
- Gsaller F, Hortschansky P, Beattie SR, Klammer V, Tuppatsch K, Lechner BE, Rietzschel N, Werner ER, Vogan AA, Chung D, Muhlenhoff U, Kato M, Cramer RA, Brakhage AA and Haas H (2014). The Janus transcription factor HapX controls fungal adaptation to both iron starvation and iron excess. EMBO J 33(19): 2261-76. doi: 10.15252/embj.201489468
- Hsu PC, Yang CY and Lan CY (2011). Candida albicans Hap43 is a repressor induced under low-iron conditions and is essential for iron- responsive transcriptional regulation and virulence. Eukaryot Cell 10(2): 207-25. doi: 10.1128/EC.00158-10
- Jung WH, Saikia S, Hu G, Wang J, Fung CK, D’Souza C, White R and Kronstad JW (2010). HapX positively and negatively regulates the transcriptional response to iron deprivation in Cryptococcus neoformans. PLoS Pathog 6(11): e1001209. doi: 10.1371/journal.ppat.1001209
- Chen C, Pande K, French SD, Tuch BB and Noble SM (2011). An iron homeostasis regulatory circuit with reciprocal roles in Candida albicans commensalism and pathogenesis. Cell Host Microbe 10(2): 118-35. doi: 10.1016/j.chom.2011.07.005
- Schrettl M, Beckmann N, Varga J, Heinekamp T, Jacobsen ID, Jochl C, Moussa TA, Wang S, Gsaller F, Blatzer M, Werner ER, Niermann WC, Brakhage AA and Haas H (2010). HapX-mediated adaption to iron starvation is crucial for virulence of Aspergillus fumigatus. PLoS Pathog 6: e1001124. doi: 10.1371/journal.ppat.1001124
- Skrahina V, Brock M, Hube B and Brunke S (2017). Candida albicans Hap43 Domains Are Required under Iron Starvation but Not Excess. Front Microbiol 8, 2388. doi: 10.3389/fmicb.2017.02388
- Mercier A, Pelletier B and Labbe S (2006). A transcription factor cascade involving Fep1 and the CCAAT-binding factor Php4 regulates gene expression in response to iron deficiency in the fission yeast Schizosaccharomyces pombe. Eukaryot Cell 5(11): 1866-81. doi: 10.1128/EC.00199-06
- Mercier A, Watt S, Bahler J and Labbe S (2008). Key function for the CCAAT-binding factor Php4 to regulate gene expression in response to iron deficiency in fission yeast. Eukaryot Cell 7(3): 493- 508. doi: 10.1128/EC.00446-07
- Mercier A and Labbe S (2009). Both Php4 function and subcellular localization are regulated by iron via a multistep mechanism involving the glutaredoxin Grx4 and the exportin Crm1. J Biol Chem 284(30): 20249-62. doi: 10.1074/jbc.M109.009563
- Lopez-Berges MS, Capilla J, Turra D, Schafferer L, Matthijs S, Jochl C, Cornelis P, Guarro J, Haas H and Di Pietro A (2012). HapX-mediated iron homeostasis is essential for rhizosphere competence and virulence of the soilborne pathogen Fusarium oxysporum. Plant Cell 24(9): 3805-22. doi: 10.1105/tpc.112.098624
- Wang Y, Deng C, Tian L, Xiong D, Tian C and Klosterman SJ (2018). The Transcription Factor VdHapX Controls Iron Homeostasis and Is Crucial for Virulence in the Vascular Pathogen Verticillium dahliae. mSphere 3(5): e00400-18. doi: 10.1128/mSphere.00400-18
- Krober A, Scherlach K, Hortschansky P, Shelest E, Staib P, Kniemeyer O and Brakhage AA (2016). HapX Mediates Iron Homeostasis in the Pathogenic Dermatophyte Arthroderma benhamiae but Is Dispensable for Virulence. PLoS One 11: e0150701. doi: 10.1371/journal.pone.0150701
- Hortschansky P, Ando E, Tuppatsch K, Arikawa H, Kobayashi T, Kato M, Haas H and Brakhage AA (2015). Deciphering the combinatorial DNA-binding code of the CCAAT-binding complex and the iron-regulatory basic region leucine zipper (bZIP) transcription factor HapX. J Biol Chem 290(10): 6058-70. doi: 10.1074/jbc.M114.628677
- Finkel JS, Xu W, Huang D, Hill EM, Desai JV, Woolford CA, Nett JE, Taff H, Norice CT, Andes DR, Lanni F and Mitchell AP (2012). Portrait of Candida albicans adherence regulators. PLoS Pathog 8(2): e1002525. doi: 10.1371/journal.ppat.1002525
- Sekonyela R, Palmer JM, Bok JW, Jain S, Berthier E, Forseth R, Schroeder F and Keller NP (2013). RsmA regulates Aspergillus fumigatus gliotoxin cluster metabolites including cyclo(L-Phe-L-Ser), a potential new diagnostic marker for invasive aspergillosis. PLoS One 8(5): e62591. doi: 10.1371/journal.pone.0062591
- Shaaban MI, Bok JW, Lauer C and Keller NP (2010). Suppressor mutagenesis identifies a velvet complex remediator of Aspergillus nidulans secondary metabolism. Eukaryot Cell 9(12): 1816-24. doi: 10.1128/EC.00189-10
- Yin WB, Amaike S, Wohlbach DJ, Gasch AP, Chiang YM, Wang CC, Bok JW, Rohlfs M and Keller NP (2012). An Aspergillus nidulans bZIP response pathway hardwired for defensive secondary metabolism operates through aflR. Mol Microbiol 83(5): 1024-34. doi: 10.1111/j.1365-2958.2012.07986.x
- Kumar NV, Yang J, Pillai JK, Rawat S, Solano C, Kumar A, Grotli M, Stemmler TL, Rosen BP and Tamas MJ (2015). Arsenic Directly Binds to and Activates the Yeast AP-1-Like Transcription Factor Yap8. Mol Cell Biol 36(6): 913-22. doi: 10.1128/MCB.00842-15
- Veide Vilg J, Kumar NV, Maciaszczyk-Dziubinska E, Sloma E, Onesime D, Aubert J, Migocka M, Wysocki R and Tamas MJ (2014). Elucidating the response of Kluyveromyces lactis to arsenite and peroxide stress and the role of the transcription factor KlYap8. Biochim Biophys Acta 1839(11): 1295-306. doi: 10.1016/j.bbagrm.2014.09.004
- Hahn JS, Neef DW and Thiele DJ (2006). A stress regulatory network for co-ordinated activation of proteasome expression mediated by yeast heat shock transcription factor. Mol Microbiol 60(1): 240-51. doi: 10.1111/j.1365-2958.2006.05097.x
ACKNOWLEDGMENTS
The authors are indebted to Professor Cecília Arraiano for critical reading the manuscript.
This work was financially supported by the Project LISBOA-01-0145-FEDER-007660 (Microbiologia Molecular, Estrutural e Celular, MOSTMICRO) funded by FED-ER funds through COMPETE2020 – Programa Operacional Competitividade e Internacionalização (POCI) given to CRP and of “Fundação para a Ciência e a Tecnologia” (FCT) (IF/00124/2015 to C.P) and was financially supported by grants to SC and to A.G.C for fellowships (036/BI/2018, SFRH/BD/118866/2016) respectively. The work of FD is fund-ed by the Agence Nationale pour la Recherche (CANDIHUB project, grant number ANR-14-CE14-0018-02 and STRUDYEV project, grant number ANR-10-JCJC-1603).
COPYRIGHT
© 2019

Yeast AP-1 like transcription factors (Yap) and stress response: a current overview by Rodrigues-Pousada et al. is licensed under a Creative Commons Attribution 4.0 International License.