Research Articles:
Microbial Cell, Vol. 5, No. 9, pp. 393 - 403; doi: 10.15698/mic2018.09.645
Single telomere length analysis in Ustilago maydis, a high-resolution tool for examining fungal telomere length distribution and C-strand 5’-end processing
1 Department of Microbiology & Immunology, W. R. Hearst Microbiology Research Center, Weill Cornell Medical College, New York, New York, United States of America.
2 Sandra and Edward Meyer Cancer Center, Weill Cornell Medical College, New York, New York, United States of America.
Keywords: telomere, telomerase, STELA, Blm, Trt1, Ustilago maydis.
Abbreviations:
STELA – single telomere length analysis,
TRF – telomere restriction fragement.
Received originally: 16/05/2018 Received in revised form: 10/07/2018
Accepted: 12/08/2018
Published: 07/08/2018
Correspondence:
Neal F. Lue, Department of Microbiology and Immunology, Weill Cornell Medical College, 1300 York Avenue, New York, NY 10065; Tel: +1 212 746-6506;Fax: +1 212 746-8587; nflue@med.cornell.edu
Conflict of interest statement: The authors declare no conflict of interests.
Please cite this article as: Ganduri Swapna, Eun Young Yu and Neal F. Lue (2018). Single telomere length analysis in Ustilago maydis, a high-resolution tool for examining fungal telomere length distribution and C-strand 5’-end processing. Microbial Cell 5(9): 393-403. doi: 10.15698/mic2018.09.645
Abstract
Telomeres play important roles in genome stability and cell proliferation. Telomere lengths are heterogeneous and because just a few abnormal telomeres are sufficient to trigger significant cellular response, it is informative to have accurate assays that reveal not only average telomere lengths, but also the distribution of the longest and shortest telomeres in a given sample. Herein we report for the first time, the development of single telomere length analysis (STELA)—a PCR-based assay that amplifies multiple, individual telomeres— for Ustilago maydis, a basidiomycete fungus. Compared to the standard telomere Southern technique, STELA revealed a broader distribution of telomere size as well as the existence of relatively short telomeres in wild type cells. When applied to blm∆, a mutant thought to be defective in telomere replication, STELA revealed preferential loss of long telomeres, whose maintenance may thus be especially dependent upon efficient replication. In comparison to blm∆, the trt1∆ (telomerase null) mutant exhibited greater erosion of short telomeres, consistent with a special role for telomerase in re-lengthening extra-short telomeres. We also used STELA to characterize the 5’ ends of telomere C-strand, and found that in U. maydis, they terminate preferentially at selected nucleotide positions within the telomere repeat. Deleting trt1 altered the 5’-end distributions, suggesting that telomerase may directly or indirectly modulate C-strand 5’ end formation. These findings illustrate the utility of STELA as well as the strengths of U. maydis as a model system for telomere research.
INTRODUCTION
Eukaryotic chromosome ends, or telomeres, play critical functions in maintaining genome stability and controlling cell proliferation. Telomere DNA comprises numerous copies of a short repeat, which is G-rich on the 3’-end-containing strand (G-strand) and C-rich on the complementary, 5’-end-containing strand (C-strand). This telomere DNA nucleates the assembly of a special nucleoprotein structure at chromosome ends, which in turn allows the cells to distinguish the normal ends from abnormal double strand breaks (DSBs) and to avoid inappropriate telomere repair [1][2]. Telomeres can display varying degrees of dysfunction. Some abnormal telomeres trigger a DNA damage response that induces growth arrest [3]. Other, more fully “deprotected” telomeres, are fusogenic and can trigger cycles of genomic re-arrangement [4][5].
–
Notably, an adequate amount of telomere DNA is required to promote the formation of a functional telomere, and different degrees of telomere loss are associated with different severities of telomere dysfunction [3]. However, despite its crucial importance, telomere DNA is difficult to maintain for two reasons. First, owing to the propensity of G-rich telomere DNA to adopt G-quadruplex (G4) or related structures, replication forks often stall in the telomere region, resulting in stochastic telomere truncation [6]. Accordingly, efficient telomere replication depends critically on multiple helicases and recombinational repair proteins (e.g., BLM, WRN, RTEL, FEN1, RAD51, BRCA2 and DNA2) that help overcome replication barriers or stabilize stalled forks [6][7][8]. Second, owing to the end replication problem, telomere DNA experiences progressive shortening following each round of replication [9]. To compensate for this loss, most eukaryotes rely on telomerase, a cellular reverse transcriptase that extends the G-strand of telomeres [10][11], and primase-Pol a, a replicative polymerase that specializes in the synthesis of the C-strand [12], thereby converting the newly generated 3’-overhangs to mostly duplex DNA. Hence, telomere dynamics in a cell population are modulated by multiple telomere-lengthening and telomere-shortening mechanisms.
–
We are particularly interested in understanding the interplay between telomere replication and telomerase in sustaining telomere length. For this purpose, we have been exploring the Basidiomycete fungus Ustilago maydis as a useful model system [13][14][15]. In comparison to the standard budding and fission yeast models, U. maydis has a number of advantages, especially in regard to the study of telomere replication. First, unlike budding and fission yeasts, U. maydis has a telomere repeat sequence (TTAGGG/CCCTAA) that is identical to the human sequence. This is attractive because the propensity of the telomere sequence to form G4-related structures is believed to underlie the special difficulty of replicating through telomere DNA. Given that different G-rich sequences may adopt different structures, the sharing of identical telomere sequence in U. maydis and mammals allows one to obviate a confounding variable. Second, we showed earlier that just like their mammalian counterparts, several U. maydis helicase and repair mutants exhibit apparent telomere replication defects. In particular, the U. maydis rad51∆, brh2∆ (~BRCA2 deletion), and blm∆ mutants all exhibit prominent telomere shortening even in telomerase-positive cells [13][14]. In contrast, even though some budding yeast helicase and repair proteins have also been implicated in telomere maintenance, this role is typically more discernable in telomerase-negative cells—e.g., Saccharomyces cerevisiae SGS1 or RAD51 deletion can accelerate senescence of telomerase-negative mutants, but does not cause telomere shortening in wild type cells [16][17]. By characterizing the U. maydis helicase/repair mutants and trt1∆ (telomerase-null) separately and in combination, we showed that the helicase/repair genes probably promote telomere maintenance through overlapping pathways that are distinct from the telomerase pathway [15]. For example, in contrast to the progressive shortening phenotype of trt1∆, telomeres are stably maintained in the helicase/repair mutants, and combining the helicase/repair mutations with trt1∆ triggers accelerated senescence and telomere loss.
–
In these previous examinations of telomere maintenance defects, we utilized Southern analysis of telomere restriction fragments (TRFs) as the assay for measuring telomere lengths. This long-standing, standard method suffers from several significant drawbacks owing to the size heterogeneity of the terminal repeat tracts, which vary by hundreds of base pairs. First, because the TRF clusters are visualized as smears in the assay, it is difficult to define accurately the upper and lower boundaries of these clusters. Second, because shorter telomere tracts yield weaker signals — as a result of binding fewer probes, the low range of the telomere distribution is prone to being underestimated. This is problematic because the telomere loss of the repair mutants may occur at only a small fraction of the telomeres, and because only a few abnormally short telomeres are sufficient to trigger a cellular response [3]. To overcome the deficiencies of TRF Southern, investigators have sought to develop alternative methods for defining telomere length distributions [18]. An example is single telomere length analysis (STELA), a PCR-based method initially developed to characterize human telomeres [19]. In this assay, the 5’-end of telomere C-strand is first ligated to an anchor oligo, allowing the modified DNA to be subsequently amplified using a pair of primers that correspond to the anchor sequence and a chromosome-specific subtelomeric sequence, respectively (Fig. 1A). Variations of STELA were later developed to allow for simultaneous measurements of telomeres from all different chromosome ends [20][21]. STELA has been particularly valuable in characterizing short telomeres, which, as noted before, are often under-identified. Interestingly, despite its potential utility, no STELA assay has been reported for either the budding or fission yeast telomeres. This is due in part to the irregular telomere repeats in both fungi, which render the design of anchor oligos challenging. While individual telomeres in S. cerevisiae have been investigated by cloning and sequencing [22], this labor-intensive strategy is not well suited to high throughput analysis of telomeres from many samples.
–
Because U. maydis has the same telomere repeat unit as mammals, the STELA protocol can be more readily applied to this fungus. Here we describe a modified STELA assay for U. maydis telomeres, which enabled us to detect individual chromosome ends bearing two types of subtelomeric repeats. Using this assay, we demonstrated a wider distribution of telomere sizes in wild type U. maydis, as well as the existence of relatively short telomeres. We also applied this assay to blm∆ and trt1∆ mutants, and found that the former exhibits a selective loss of long telomeres, whereas the latter manifests comparable erosion of both long and short telomeres. Moreover, we showed that the 5’ ends of U. maydis telomere C-strands preferentially terminate at several positions within the repeat, and these positions differ from that previously reported for mammals. Deleting trt1 but not blm altered the distribution of telomere 5’ ends. These findings demonstrate the potential of the U. maydis STELA technique to provide new insights on telomere regulation.
RESULTS
STELA allows for measurements of individual telomere lengths in U. maydis
The STELA assay entails PCR amplification of individual telomeric fragments through the use of an anchor primer as well as a subtelomeric primer. Previous analysis of U. maydis subtelomeres revealed two classes of elements, named UTASa and UTASb. Individual members of UTASa include UT4 and UT5, whereas members of UTASb include UT6, UT7 and UT8 [23]. To generate STELA fragments from U. maydis, we first tested a subtelomeric primer that bears a sequence shared by the UT4 and UT5 subtelomeres, and that is located ~650 bp proximal to the TTAGGG terminal repeats (Fig. 1A) [24]. Following PCR and Southern analysis using a UT4/5 probe, fragments that range in size from ~800 to 1,500 bp (average of 1,150 bp) were detected (Fig. 1B). Notably, in a parallel telomere restriction fragment (TRF) analysis, the same probe identified fragments with an average size of ~1.4 kb among Bam HI-digested genomic DNA (Fig 1B). Because a Bam HI site is located ~300 bp proximal to the UT4/5 forward primer, the average sizes of the STELA and TRF DNA are in good agreement with each other. Moreover, as expected, identical fragments were detected by the UT4/5 probe and the (TTAGGG)82 probe in a sequential hybridization experiment (Fig. 1C). Notably, the STELA reactions presented in Fig. 1C utilized higher amount of template DNA than reactions in Fig. 1B, leading to higher number of products and more similar size distributions for the different reactions. Also notably, relative to previous estimates [13], STELA revealed a slightly longer average telomere length (~500 bp) and greater telomere length heterogeneity (~150 to 850 bp, i.e., a maximal variation of 700 bp) for wild type U. maydis (Fig. 1C). These previous studies, unlike the TRF analysis in Fig. 1B, were based on probing PstI-generated TRF clusters using a telomere repeat (TTAGGG) probe, and estimated the average telomere lengths to be 300-400 bp or 400 bp [13][15]. One of the studies also yielded a telomere size variation of ~ 300 bp [13]. The discrepancies between these estimates from the current estimates are not surprising given the significant uncertainty involved in TRF-based telomere length analysis. In particular, it is difficult to judge the upper and lower boundaries of a TRF cluster (see e.g., the Southern analysis in Fig. 1B). The challenge becomes even more significant when there are multiple, overlapping TRF clusters, as was the case in the earlier studies [13][15].
–
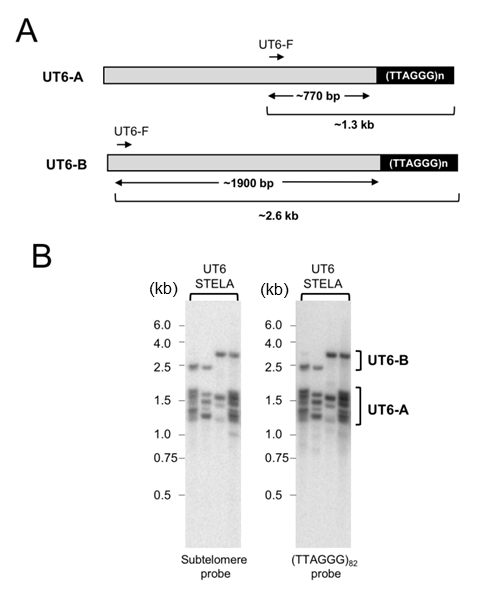
To assess the generality of our initial findings, we utilized a different subtelomeric primer with a UT6-specific sequence (UT6-F) (Fig. 2A and 2B). Based on the literature, this primer is 519 bp proximal to the terminal repeats [23]. However, as discussed below, cloning and sequencing of the STELA products suggests that the UT6 subtelomeres are longer (~770 bp) in our strain. Interestingly, probing the STELA products with either a UT6-probe or the (TTAGGG)82 probe revealed two clusters of products (designated as UT6-A and UT6-B in Fig. 2). Notably, the UT6-A products are ~ 5-fold more abundant than the UT6-B products. These observations suggest that in addition to the previously characterized UT6 element, there is a second, rarer type of UT6-containing telomeres in which the UT6-F primer is about 1.9 kb away from the terminal repeats.
–
To further confirm the accuracy of our deduction with regard to the sizes of the UT4 and UT6 subtelomere lengths in the STELA fragments, we cloned and sequenced a number of the PCR products (Supp. Fig. 1 and Supp. Fig. 2). Interestingly, we detected small sequence differences between all five UT4/5 and all five UT6 clones. For the UT4/5 clones, the sizes of the subtelomere segments range from 612 to 658 bp, close to that expected from a previous report (i.e., 612 bp [23]). However, for the UT6 clones, the subtelomere segments are substantially longer than expected and range from 734 to 805 bp. While unexpected, this observation is consistent with our finding that the UT6-A STELA products are on average ~150 bp longer than the UT4 products (1.3 kb vs 1.15 kb). Notably, none of our UT6 clones harbor an ~1.9 kb subtelomere segment predicted for UT6-B, consistent with the lower abundance of this type of subtelomeric elements.
–
The Blm helicase is critical for the maintenance of long, but not short telomere tracts
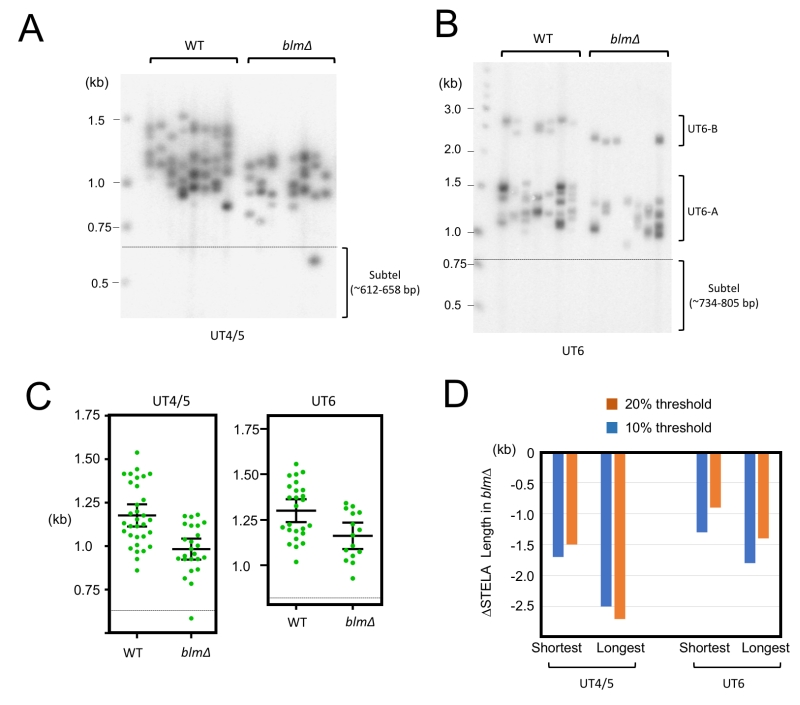
The Blm helicase has been suggested to promote telomere replication by unwinding G-quadruplexes and related structures that form within the G-rich telomere repeats [25][26]. We have previously observed significant telomere loss in the U. maydis blm∆ mutant, and estimated the average shortening to be ~220 bp [15]. By subjecting blm∆ to STELA, we observed a striking defect of the mutant in the maintenance of long telomeres (Fig. 3). For example, in the UT4/5 assays, the blm∆ STELA samples did not contain any fragments in the 1.2—1.5 kb range. Such fragments, which contain ~550 to 850 bp of repeats, constitute close to half of the products in wild type samples (Fig. 3A and 3C). In contrast, the shortest UT4/5 STELA bands in both wild type and blm∆ samples are ~750-800 bp (corresponding to ~100-150 bp of telomeres). The complete absence of very long telomeres was also observed in the UT6 STELA assays for blm∆ (Fig. 3B and 3C).
–
To quantify the effects of blm deletion on the maintenance of long and short telomeres, we determined the threshold STELA lengths for the upper and lower 10% and 20% of the products, and then calculated the changes in these values in blm∆ relative to the parental control (Fig. 3D). Consistent with preferential loss of long telomeres, we found that the upper threshold lengths for blm∆ exhibited more erosion than the lower threshold lengths (e.g., ~250 bp vs ~150 bp for the UT4 products). Because current evidence points to a role for Blm in promoting telomere replication, our findings suggest that efficient replication is most important for the maintenance of long telomeres. Notably, this observation does not imply a special replication mechanism or a special Blm function for the most distal portion of the telomeres (see Discussion).
–
Telomerase is needed to maintain minimal telomere lengths
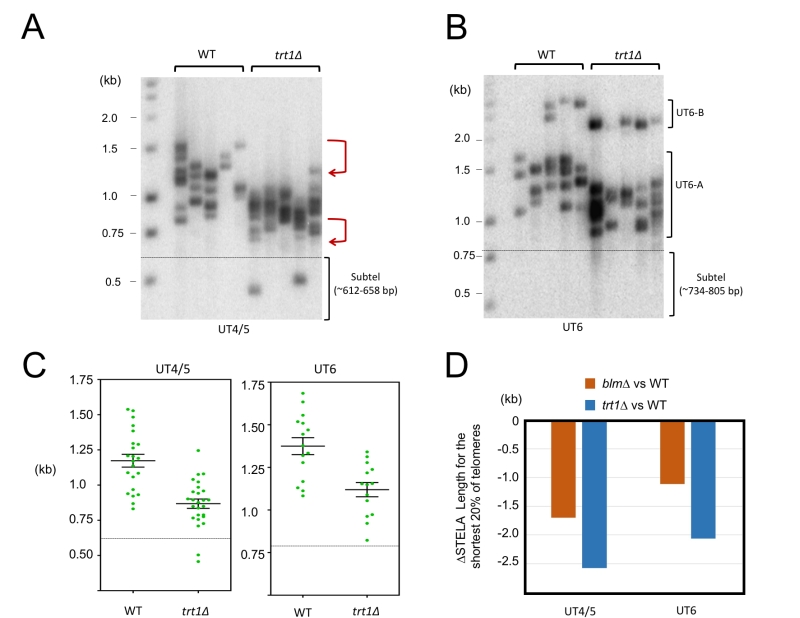
The preferential loss of long telomeres in blm∆ suggests the existence of compensatory mechanisms that allow for the maintenance of short but not long telomeres. Indeed, there is substantial evidence that telomerase preferentially elongates short telomeres [22]. We therefore analyzed telomere distributions in the trt1∆ mutants by STELA (Fig. 4A, 4B and 4C). To minimize complications introduced by post senescent survivors, we isolated DNA from trt1∆ ~100 generations after gene deletion—senescence typically occurs at 200 generation. Compared to wild type telomeres, trt1∆ telomeres exhibit evident length reduction in both the upper and lower range of the size distribution (marked by two bent arrows in Fig. 4A). This is in contrast to blm∆, which manifests a more prominent reduction in the upper range. To confirm this visual impression, we calculated the average length for the shortest 20% of telomeres in the two mutants and compared them to that in the wild type strain (Fig. 4D). As predicted, the trt1∆ mutant suffered ~2-fold more telomere contraction than blm∆, supporting the notion that telomerase plays an important role in preventing the further erosion of short telomeres.
–
U. maydis telomere C-strands terminate at preferred nucleotide positions within the 6-nt repeat unit and this preference is altered in trt1∆
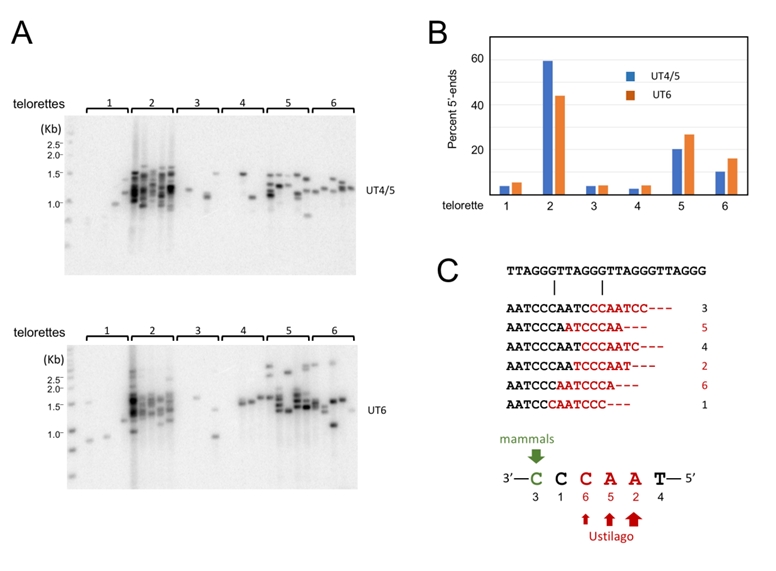
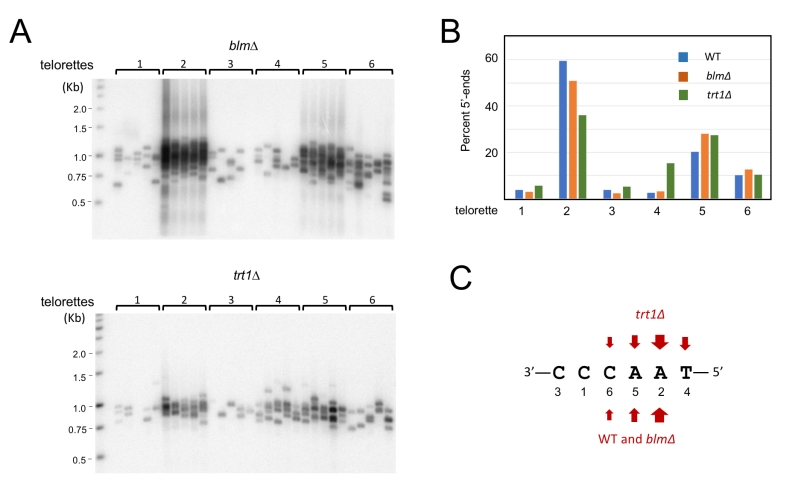
In addition to revealing detailed telomere length distribution, STELA can be used to characterize the precise 5’-ends of the telomere C-strand because the ligation step requires precise juxtaposition of the 3’-end of telorette oligo and the 5’-end of C-strand [19][27]. Through STELA analysis, mammalian telomere C-strands were previously shown to terminate at a preferred position with the telomere repeat, suggesting that the formation of the 5’ ends is subject to regulation [27][28]. No comparable studies have been reported for any fungal telomeres. We therefore characterized the U. maydis telomere C-strand termini using individual telorette oligos (Fig. 5A). Interestingly, three of the six telorette oligos (2, 5, and 6) supported the amplification of substantially higher number of STELA products, suggesting that they define the preferred 5’-ends of C-strands. This observation applies to both UT4/5 and UT6 amplifications of ligated DNA (Fig. 5A). One of the three preferred ends, 5’-AACCCT (telorette 2), accounts for ~50-60% of the termini. The other two preferred ends, 5’-ACCCTA and 5’-CCCTAA, each accounts for ~10 to 20% of the termini (Fig. 5B). Notably, the three preferred 5’-ends are clustered together, suggesting that one underlying mechanism may be responsible. Also notably, these preferred 5’-ends for U. maydis C-strand are different from that defined for mammalian telomere, which is 5’-CTAACC [27] (Fig. 5C). Thus, while preferential termination at a specific position(s) is a conserved property of telomere C-strand, the identity of the preferred position is not conserved between fungi and mammals.
–
Next, we examined the impact of deleting blm or trt1 on the distributions of C-strand 5’ ends on UT4/5 telomeres (Fig. 6). In comparison to the parental strain, only the trt1∆ mutant exhibited significant differences in its 5’ end distribution: the most predominant end in the parental strain (5’-AACCCT; telorette 2) became less well represented, and a rarely used end (5’-TAACCC; telorette 4) became more abundant (Fig. 6B and 6C). Most notably, the frequency of telorette 4 utilization is elevated from 2.5% in wild type DNA to 15.4% in trt1∆ DNA, representing a 6-fold increase. The observed alterations in the trt1∆ telomere 5’-ends were confirmed in three additional sets of STELA assays for the wild type and trt1∆ DNA; both the reduction in telorette 2 products and the increase in telorette 4 products in trt1∆ DNA were reproducible and statistically significant (Supp. Fig. 3). This result suggests that telomerase can directly or indirectly modulate C-strand 5’ end formation.
DISCUSSION
In this study, we developed the first STELA assay for a fungus, and used it to characterize the telomere lengths and telomere 5’-end structures of this fungus. Our preliminary utilization of this assay to characterize the telomeres in the wild type and two mutant strains revealed a number of interesting findings as well as discrepancies from previous studies. The implications of these observations are discussed below.
–
Comparison of the STELA results with previous characterizations of telomere lengths and subtelomeric structures
The size range of telomere repeat tracts as determined by STELA (a maximal variation of 700 bp) is significantly broader than that by Southern analysis (e.g., a maximal variation of about 300 bp in one study [15]). This is not surprising given the limited resolution of the TRF analysis, and the uncertainty in marking the upper and lower boundaries of a TRF cluster. Moreover, because a short telomere yields weaker hybridization signal in Southern analysis, its presence is more likely to go undetected. In light of these considerations, it is notable that we found the minimal telomeres in the wild type U. maydis strain to be ~ 150 bps, suggesting that at this length, telomeres are recognized as sufficiently long to avoid obligatory extension by telomerase. STELA thus allows us to gain a better estimate of how telomerase activity is regulated by the length of telomeres.
–
One caveat of the STELA analysis is that it relies on PCR amplification to identify individual telomeres, and a priori, the PCR efficiency of individual telomeres may differ. However, two observations suggest that this is not a significant issue. First, in contrast to mammalian STELA fragments, which often vary by 5-10 kb in length in a given reaction, the U. maydis fragments are much more homogeneous in size. Second, most of the U. maydis STELA products in a given experiment have similar intensities, as would be predicted if they are amplified with similar efficiency. Importantly, there is no evidence that the shorter telomeres are more efficiently amplified; even the few STELA fragments that are evidently devoid of telomeric repeats (e.g., the blm∆ band below the dashed line in Fig. 3A) are amplified to the same level. Thus, the protocol we have developed appears to amplify the UT-4/5 and UT-6 telomeres in an unbiased manner, providing a fair representation of the total telomere population.
–
Previous analysis of U. maydis subtelomeres revealed two classes of elements, named UTASa (including UT4 and UT5) and UTASb (including UT6, UT7 and UT8). In our STELA analysis, we utilized two forward primers, one shared by UT4 and UT5, and the other specific to UT6 [23]. Cloning of the UT4/5 primer-derived products confirmed that these elements are highly similar to one another, and revealed additional variations in the length and sequences of these related elements. In particular, the sizes of the subtelomeric fragments as de-limited by the STELA forward primer and the TTAGGG repeats vary by ~ 50 bp. Such variations are also observed in the UT6 primer-derived products, which differ in length by as much as 70 bp. Moreover, we identified a rare variant subtelomere in which the UT6 primer is positioned about 1.9 kb away from the terminal repeat tract. Based on the relative abundance of STELA products, this rare variant is present at ~ 1/5 the frequency of the typical UT6 element. By further characterizing this and other rare variants, we may in the future be able to perform chromosome-specific STELA and characterize inter-chromosomal differences in telomere structure and dynamics, similar to what has been done for human telomeres [29].
–
The distinct mechanisms of Blm and telomerase in promoting telomere maintenance
A somewhat unique strength of U. maydis as a model system for telomere research is that like mammalian cells, mutations in U. maydis genes that promote telomere replication (e.g., rad51, blm) cause telomere loss in telomerase-positive cells [13][15]. Such phenotypes have not been reported in comparable budding and fission yeast mutants. We have previously proposed that the telomere loss of these U. maydis mutants could be explained by stochastic truncation of telomeres due to fork stalling/collapse followed by incomplete re-extension by telomerase [15]. The current analysis provides some additional insights on this balance. Most notably, the blm∆ mutant exhibits a dramatic and selective loss of long telomeres; telomeres with 550-850 bp repeat tracts, which are abundant in the parental strain, are completely missing in blm∆. This result suggests that complete replication of a telomere tract more than 550 bp long is problematic in the absence of Blm. In contrast, the minimal telomere length in the blm∆ mutant is similar to that of the parent strain. For example, in the UT4/5 STELA analysis, only 3 out 24 STELA fragments in the blm∆ sample have telomeres that are shorter than 150 bp (Fig. 3A and 3C), the minimal length found in the parental strain. The loss of very long telomeres in blm∆ and the retention of the 150 bp minimal telomere tracts do not necessarily imply a special Blm mechanism in the most distal portion of telomeres. First, it is possible that during replication, short telomere tracts are subjected to less frequent G4-related blockade, and hence less dependent on Blm for fork progression. Alternatively, telomerase may efficiently elongate very short telomeres and prevent their accumulation. Consistent with the second mechanism, when trt1 is deleted, there is a substantial increase in the abundance of 50-150 bp telomeres, even in early passages (Fig. 4).
–
Telomere C-strand maturation in U. maydis
A key advantage of the STELA assay is that it allows for the precise determination of telomere C-strand 5’ ends at individual chromosome termini. Previous examination of mammalian telomeres revealed the predominance of a specific 5’ end sequence (i.e., 5’-CTAACC), indicative of specific processing steps that favor the removal of more distal ribonucleotides and deoxyribonucleotides generated by primase-Pol a during C-strand synthesis [27][28]. We performed a comparable analysis of U. maydis telomeres, and identified three preferred 5’ ends, which together account for ~80% of chromosome ends (Fig. 5). Interestingly, none of these ends correspond to the preferred mammalian telomere 5’ end. Thus, while preferential termination at a specific position(s) is a conserved property of telomere C-strand, the identity of the preferred position is not shared between fungi and mammals, even for fungi that bear the canonical telomere repeat sequence. This finding suggests that either the processing nucleases in these organisms have different properties, or that they are regulated differently by the telomere nucleoprotein structure.
–
It is notable that deleting trt1 altered the distribution of the U. maydis C-strand 5’ end nucleotides. This is somewhat unexpected given that in the previous mammalian study, telomerase-positive and -negative cells were found to exhibit the same 5’-end preference [27]. Since telomerase is not thought to be directly involved in C-strand formation, this effect in U. maydis may be mediated through an indirect mechanism. For example, it has been shown that in G-strand elongation, telomerase preferentially pause or terminate at the GGTTAG position, which is complementary to the last RNA template residue. This preferred G-strand terminus could indirectly influence the positioning of the last C-strand RNA-DNA chimera in the next cell cycle, or influence the positioning of G-strand-binding proteins such as Pot1. These positioning preferences could in turn modulate the eventual 5’ end of the C-strand. When telomerase is absent, the G-strand 3’ ends may become more heterogeneous, resulting in correspondingly greater heterogeneity in the C-strand 5’ ends. Additional studies will be required to test this and other interesting possibilities.
–
In summary, we have developed a STELA assay for U. maydis, and demonstrated the potential of this assay for characterizing telomere length distribution as well as the mechanisms of C-strand 5’ end formation. This represents the first STELA-based analysis for fungal telomeres, and provides further illustrations of the utility and versatility of U. maydis as a model system for investigating telomere mechanisms.
MATERIALS AND METHODS
Ustilago maydis strains and growth conditions
Standard protocols were employed for the genetic manipulation of U. maydis [30][31][32]. All U. maydis strains used in this study were haploid and were derived from the UCM350 background [31][33]. These strains have all been described before and are listed in Supp. Table 1.
–
Southern analysis of telomere restriction fragments
Southern analysis of telomere restriction fragments (TRF) was performed using DNA treated with BamHI [13]. The blot was hybridized to a labeled subtelomeric fragment generated by PCR using the UT4/5-F and UT4-subtel-R2375 primers (Supp. Table 2).
–
STELA
DNA from U. maydis strains was extracted using lysing enzyme and the GeneJET genomic DNA purification kit (Thermo Fisher Scientific). Ligation to telorette oligos (Supp. Table 2 and [27]) were performed in 15 µl reactions containing 1 ng genomic DNA, 0.001 µM telorette oligo, 1x CutSmart Buffer (NEB), 1 mM ATP, and 800 U T4 DNA ligase (NEB) at 35°C for 20 hours. PCR assays were carried out in 25 µl reactions containing 2.5¾10 pg template DNA, 1 µM each of the subtelomeric forward primer (UT4-F or UT6-F) and the teltail reverse primer, 1x Failsafe PCR PreMix H, and 2.5 U of Failsafe polymerase (Lucigen). Cycling conditions were as follows: 30 sec at 94°C, 30 sec at 63°C and 2 min at 72°C. 33 and 35 cycles were utilized for the UT4/5-telomere and UT6-telomere PCR reactions, respectively. To ensure adequate coverage of the telomere size distribution, we performed 4-8 parallel PCR reactions for each ligated DNA sample. The reaction products were analyzed by electrophoresis in 0.9 % agarose gels and subjected to Southern using the appropriate subtelomeric probes or a telomere repeat probe ((TTAGGG)82). Following PhosphorImager scanning (GE Healthcare), the sizes of individual telomeres were determined using TESLA software [20], and the results analyzed and plotted using Prism (GraphPad Software).
–
For cloning of STELA products containing UT4/5 and UT6 sequences, we increased the cycle numbers to 40 and 50, respectively. Following PCR amplification, the DNA was isolated using the Monarch® PCR & DNA Cleanup Kit (NEB, Inc.) and then introduced into the pMiniT 2.0 vector using the NEB® PCR Cloning Kit. The inserts were sequenced using the forward or reverse primer provided by the kit (pMiniT 2.0 forward and pMiniT 2.0 reverse, see Supp. Table 2).
References
- T. de Lange, "How Telomeres Solve the End-Protection Problem", Science, vol. 326, pp. 948-952, 2009. http://dx.doi.org/10.1126/science.1170633
- N. Arnoult, and J. Karlseder, "Complex interactions between the DNA-damage response and mammalian telomeres", Nature Structural & Molecular Biology, vol. 22, pp. 859-866, 2015. http://dx.doi.org/10.1038/nsmb.3092
- Z. Kaul, A.J. Cesare, L.I. Huschtscha, A.A. Neumann, and R.R. Reddel, "Five dysfunctional telomeres predict onset of senescence in human cells", EMBO reports, vol. 13, pp. 52-59, 2011. http://dx.doi.org/10.1038/embor.2011.227
- R. Capper, B. Britt-Compton, M. Tankimanova, J. Rowson, B. Letsolo, S. Man, M. Haughton, and D.M. Baird, "The nature of telomere fusion and a definition of the critical telomere length in human cells", Genes & Development, vol. 21, pp. 2495-2508, 2007. http://dx.doi.org/10.1101/gad.439107
- F.D.D. Fagagna, P.M. Reaper, L. Clay-Farrace, H. Fiegler, P. Carr, T. von Zglinicki, G. Saretzki, N.P. Carter, and S.P. Jackson, "A DNA damage checkpoint response in telomere-initiated senescence", Nature, vol. 426, pp. 194-198, 2003. http://dx.doi.org/10.1038/nature02118
- P. Martínez, and M.A. Blasco, "Replicating through telomeres: a means to an end", Trends in Biochemical Sciences, vol. 40, pp. 504-515, 2015. http://dx.doi.org/10.1016/j.tibs.2015.06.003
- S. Badie, J.M. Escandell, P. Bouwman, A.R. Carlos, M. Thanasoula, M.M. Gallardo, A. Suram, I. Jaco, J. Benitez, U. Herbig, M.A. Blasco, J. Jonkers, and M. Tarsounas, "BRCA2 acts as a RAD51 loader to facilitate telomere replication and capping", Nature Structural & Molecular Biology, vol. 17, pp. 1461-1469, 2010. http://dx.doi.org/10.1038/nsmb.1943
- J. Vannier, V. Pavicic-Kaltenbrunner, M. Petalcorin, H. Ding, and S. Boulton, "RTEL1 Dismantles T Loops and Counteracts Telomeric G4-DNA to Maintain Telomere Integrity", Cell, vol. 149, pp. 795-806, 2012. http://dx.doi.org/10.1016/j.cell.2012.03.030
- C.B. Harley, A.B. Futcher, and C.W. Greider, "Telomeres shorten during ageing of human fibroblasts", Nature, vol. 345, pp. 458-460, 1990. http://dx.doi.org/10.1038/345458a0
- C. Autexier, and N.F. Lue, "The Structure and Function of Telomerase Reverse Transcriptase", Annual Review of Biochemistry, vol. 75, pp. 493-517, 2006. http://dx.doi.org/10.1146/annurev.biochem.75.103004.142412
- R.A. Wu, H.E. Upton, J.M. Vogan, and K. Collins, "Telomerase Mechanism of Telomere Synthesis", Annual Review of Biochemistry, vol. 86, pp. 439-460, 2017. http://dx.doi.org/10.1146/annurev-biochem-061516-045019
- N.F. Lue, J. Chan, W.E. Wright, and J. Hurwitz, "The CDC13-STN1-TEN1 complex stimulates Pol α activity by promoting RNA priming and primase-to-polymerase switch", Nature Communications, vol. 5, 2014. http://dx.doi.org/10.1038/ncomms6762
- E.Y. Yu, M. Kojic, W.K. Holloman, and N.F. Lue, "Brh2 and Rad51 promote telomere maintenance in Ustilago maydis, a new model system of DNA repair proteins at telomeres", DNA Repair, vol. 12, pp. 472-479, 2013. http://dx.doi.org/10.1016/j.dnarep.2013.04.027
- E.Y. Yu, J. Pérez-Martín, W.K. Holloman, and N.F. Lue, "Mre11 and Blm-Dependent Formation of ALT-Like Telomeres in Ku-Deficient Ustilago maydis", PLOS Genetics, vol. 11, pp. e1005570, 2015. http://dx.doi.org/10.1371/journal.pgen.1005570
- E.Y. Yu, M. Hsu, W.K. Holloman, and N.F. Lue, "Contributions of recombination and repair proteins to telomere maintenance in telomerase‐positive and negative Ustilago maydis", Molecular Microbiology, vol. 107, pp. 81-93, 2017. http://dx.doi.org/10.1111/mmi.13866
- F. Johnson, "The Saccharomyces cerevisiae WRN homolog Sgs1p participates in telomere maintenance in cells lacking telomerase", The EMBO Journal, vol. 20, pp. 905-913, 2001. http://dx.doi.org/10.1093/emboj/20.4.905
- B.J. Ballew, and V. Lundblad, "Multiple genetic pathways regulate replicative senescence in telomerase‐deficient yeast", Aging Cell, vol. 12, pp. 719-727, 2013. http://dx.doi.org/10.1111/acel.12099
- E. Vera, and M.A. Blasco, "Beyond average: potential for measurement of short telomeres", Aging, vol. 4, pp. 379-392, 2012. http://dx.doi.org/10.18632/aging.100462
- D.M. Baird, J. Rowson, D. Wynford-Thomas, and D. Kipling, "Extensive allelic variation and ultrashort telomeres in senescent human cells", Nature Genetics, vol. 33, pp. 203-207, 2003. http://dx.doi.org/10.1038/ng1084
- T. Lai, N. Zhang, J. Noh, I. Mender, E. Tedone, E. Huang, W.E. Wright, G. Danuser, and J.W. Shay, "A method for measuring the distribution of the shortest telomeres in cells and tissues", Nature Communications, vol. 8, 2017. http://dx.doi.org/10.1038/s41467-017-01291-z
- L. Bendix, P.B. Horn, U.B. Jensen, I. Rubelj, and S. Kolvraa, "The load of short telomeres, estimated by a new method, Universal STELA, correlates with number of senescent cells", Aging Cell, vol. 9, pp. 383-397, 2010. http://dx.doi.org/10.1111/j.1474-9726.2010.00568.x
- M. Teixeira, M. Arneric, P. Sperisen, and J. Lingner, "Telomere Length Homeostasis Is Achieved via a Switch between Telomerase- Extendible and -Nonextendible States", Cell, vol. 117, pp. 323-335, 2004. http://dx.doi.org/10.1016/S0092-8674(04)00334-4
- P. Sánchez-Alonso, and P. Guzmán, "Organization of chromosome ends in Ustilago maydis. RecQ-like helicase motifs at telomeric regions.", Genetics, 1998. http://www.ncbi.nlm.nih.gov/pubmed/9539423
- P.A. Guzmán, and J.G. Sánchez, "Characterization of telomeric regions from Ustilago maydis.", Microbiology (Reading, England), 1994. http://www.ncbi.nlm.nih.gov/pubmed/8012578
- C. Barefield, and J. Karlseder, "The BLM helicase contributes to telomere maintenance through processing of late-replicating intermediate structures", Nucleic Acids Research, vol. 40, pp. 7358-7367, 2012. http://dx.doi.org/10.1093/nar/gks407
- M. Zimmermann, T. Kibe, S. Kabir, and T. de Lange, "TRF1 negotiates TTAGGG repeat-associated replication problems by recruiting the BLM helicase and the TPP1/POT1 repressor of ATR signaling", Genes & Development, vol. 28, pp. 2477-2491, 2014. http://dx.doi.org/10.1101/gad.251611.114
- A.J. Sfeir, W. Chai, J.W. Shay, and W.E. Wright, "Telomere-End Processing", Molecular Cell, vol. 18, pp. 131-138, 2005. http://dx.doi.org/10.1016/j.molcel.2005.02.035
- T.T. Chow, Y. Zhao, S.S. Mak, J.W. Shay, and W.E. Wright, "Early and late steps in telomere overhang processing in normal human cells: the position of the final RNA primer drives telomere shortening", Genes & Development, vol. 26, pp. 1167-1178, 2012. http://dx.doi.org/10.1101/gad.187211.112
- B. Britt-Compton, J. Rowson, M. Locke, I. Mackenzie, D. Kipling, and D.M. Baird, "Structural stability and chromosome-specific telomere length is governed by cis-acting determinants in humans", Human Molecular Genetics, vol. 15, pp. 725-733, 2006. http://dx.doi.org/10.1093/hmg/ddi486
- J. Kämper, "A PCR-based system for highly efficient generation of gene replacement mutants in Ustilago maydis", Molecular Genetics and Genomics, vol. 271, pp. 103-110, 2003. http://dx.doi.org/10.1007/s00438-003-0962-8
- M. Kojic, C.F. Kostrub, A.R. Buchman, and W.K. Holloman, "BRCA2 Homolog Required for Proficiency in DNA Repair, Recombination, and Genome Stability in Ustilago maydis", Molecular Cell, vol. 10, pp. 683-691, 2002. http://dx.doi.org/10.1016/s1097-2765(02)00632-9
- A. Brachmann, J. König, C. Julius, and M. Feldbrügge, "A reverse genetic approach for generating gene replacement mutants in Ustilago maydis", Molecular Genetics and Genomics, vol. 272, pp. 216-226, 2004. http://dx.doi.org/10.1007/s00438-004-1047-z
- F. Banuett, and I. Herskowitz, "Different a alleles of Ustilago maydis are necessary for maintenance of filamentous growth but not for meiosis.", Proceedings of the National Academy of Sciences of the United States of America, 1989. http://www.ncbi.nlm.nih.gov/pubmed/16594058
SUPPLEMENTAL INFORMATION
![]() Download Supplemental Information
Download Supplemental Information
ACKNOWLEDGMENTS
We thank members of our laboratories for discussion and Bill Holloman (Weill Cornell Medicine) for strains, advice and comments on the manuscript. This work was supported by a pilot project grant from the Sandra and Edward Meyer Cancer Center, the Bohmfalk Charitable Trust, and NSF (MCB-1817331).
COPYRIGHT
© 2018

Single telomere length analysis in Ustilago maydis, a high-resolution tool for examining fungal telomere length distribution and C-strand 5’-end processing by Swapna et al. is licensed under a Creative Commons Attribution 4.0 International License.