News and thoughts:
Microbial Cell, Vol. 2, No. 1, pp. 1 - 4; doi: 10.15698/mic2015.01.185
The emerging role of complex modifications of tRNALysUUU in signaling pathways
1 Genetics and Genomics Graduate Program, University of Florida, Gainesville, Florida, USA.
2 University of Florida Genetics Institute, Gainesville, Florida, USA.
3 Institut de Génétique et Microbiologie, Université of Paris-Sud, Orsay, France.
4 Department of Microbiology and Cell Science, University of Florida, Gainesville, Florida, USA.
Keywords: tRNA, GCN4, thiolation, UPR, TOR.
Abbreviations:
mcm5s2U, 5-methoxycarbonylmethyl-2-thiouridine;
t6A , N6-threonylcarbamoyladenosine;
ASL, Anticodon-Stem-Loop;
ELP, Elongator Complex;
ER, Endoplasmic Reticulum;
GAAC, General Amino Acid Control;
KEOPS, Kinase, putative Endopeptidase and Other Proteins of Small size;
TOR, Target of Rapamycin;
UPR, Unfolded Protein Response.
Received originally: 24/12/2014 Accepted: 29/12/2014
Published: 05/01/2015
Correspondence:
Valérie de Crécy-Lagard, University of Florida Genetics Institute, Gainesville, Florida, USA vcrecy@ufl.edu
Conflict of interest statement:
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Please cite this article as: Patrick C. Thiaville and Valérie de Crécy-Lagard (2015). The emerging role of complex modifications of tRNALysUUU in signaling pathways. Microbial Cell 2(1): 1-4.
Coordination of cell growth with nutrient availability, in particular amino acids, is a central problem that has been solved by the implementation of complex regulatory cascades. Although the specific regulatory mechanisms differ between kingdoms and species, a common theme is the use of tRNA molecules as sensors and transducers of amino acid starvation. In many bacteria, amino acid starvation leads to high levels of uncharged tRNAs, a signal for the synthesis of the stringent response’s alarmones, halting transcription of stable RNAs and inducing the synthesis of amino acid synthesis pathways [1]. In gram-positive Bacteria (as well as the Deinococcus-Thermus clade), uncharged tRNAs bind structures (T-boxes) in the leader sequences of mRNA encoding gene, activating the expression of genes involved in amino acid metabolism [2]. In eukaryotes, the conserved General Amino Acid Control (GAAC) response is triggered by shortage of amino acids that leads to the binding of uncharged tRNAs to Gcn2 kinase and, through a cascade of events, to the activation of the central activator of amino acid synthesis genes, Gcn4 [3]. As the study by Scheidt et al. [4] and several other recent studies in this field reveal, variations in charging levels are not the only mechanism by which tRNAs play a role in amino acid starvation responses; levels of post-transcriptional modifications also seem to play major roles.
–
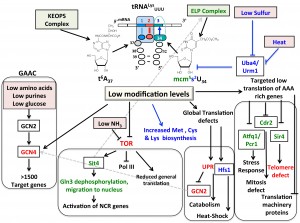
The anticodon-stem-loop (ASL) of tRNAs drives decoding by interacting directly with the mRNA codon (Fig. 1). Modifications of the ASL are the most distinct and chemically complex of all RNA modifications [5]. They are required for accurate codon recognition and translocation, enhance aminoacylation properties of tRNAs, and prevent ribosomal frameshifting [5]. In eukaryotes, two of the most complex ASL modifications are N6-threonylcarbamoyladenosine (t6A) and 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U). The first modification is found at position 37 of most tRNAs decoding ANN codons, just before the residue that interacts with the first base of the codon. The second modification is found at position 34, the residue that decodes the 3rd base of the codon, of tRNALysUUU, tRNAGluUUC, and tRNAGlnUUG (Fig. 1). Both these modifications are found in tRNALysUUU and affect translation efficiency [5]. In addition, several other striking parallels can be made between the two.
t6A and mcm5s2U are both synthesized by elaborate multimeric complexes that were first thought to be transcription factors. The KEOPS complex (Kinase, putative Endopeptidase and Other Proteins of Small size) in combination with Sua5 is involved in the synthesis of t6A [6], and the Elongator (ELP) complex in combination with the URM1 pathway enzymes (Uba4, Urm1, Ncs2/Ncs6) is involved in the synthesis of mcm5s2U [5]. Saccharomyces cerevisiae mutants lacking t6A or mcm5s2U display many similar phenotypes. For example, telomere shortening is observed in the absence of either modification [7][8]. In the case of ELP, it is now firmly established that most of the ELP phenotypes are due to the absence of the modified base, as the phenotypes are suppressed by overexpressing the tRNA targets [7][8][9]. For example, the levels of the Sir4, a regulator involved in telomere maintenance and enriched in AAA (Lys) codons, are decreased in mcm5s2U deficient strains. Overexpression of tRNALysUUU suppresses the telomere shortening phenotype [8]. In Schizosaccharomyces pombe, key regulators, including the two subunits Atf1 and Pcr1 of the central regulator of the core environmental stress response [10] and the cell division regulator Cdr2 [11], have an over-representation of AAA codons over other Lys codons, and their efficient translation is dependent on mcm5s2U levels. More generally, proteins involved in translation initiation and elongation are enriched in AAA codons, and reduced in cells lacking mcm5s2U [12][13]. It is not yet known if the levels of these proteins enriched in AAA codons also require t6A for efficient translation in yeast, but it has been shown in mammals that a decrease in sulfur modified form of t6A (ms2t6A) on tRNALysUUU leads to lower levels of proinsulin [14].
–
Another phenotype shared by a t6A or a mcm5s2U deficient S. cerevisiae derivatives is the GCN2 independent activation of GCN4 [15][16]. As the activation of the GAAC leads to a major reprograming of transcription (>500 genes are induced and >1000 are repressed) [17], this makes it difficult to interpret whether specific phenotypes observed in these tRNA modification mutants are due to direct or indirect effects. This is made all the more problematic given that another central regulator, the Target of Rapamycin (or TOR), is also affected by both t6A and mcm5s2U.
–
The TOR kinases regulate the balance between protein synthesis and protein degradation in response to nutrient quality and TOR activity is inhibited by low nitrogen conditions, caffeine or rapamycin [18]. In yeast, reduced TOR levels increase levels of the Sit4 phosphatase, which dephosphorylates TOR targets such as the regulator Gln3. Unphosporylated Gln3 will then relocate to the nucleus to activate genes required for growth on low quality nitrogen sources. The following links between TOR and mcm5s2U have previously been made: 1) ELP mutants are sensitive to rapamycin and caffeine [16]; 2) deletion of SIT4 leads to rapamycin hypersensitivity and resistance to zymocin (a tRNase that recognizes mcm5s2U and cleaves tRNALysUUU leading to cell death) [19] because Sit4 activates the ELP complex by phosphorylation [20]. As shown by Scheidt et al. [4], the Elongator mutants mislocate Gln3 leading to rapamycin hypersensitivity, a phenotype that can be suppressed by over-expression of the tRNAs modified by mcm5s2U, suggesting that proper tRNA modification affects Gln3 localization and subsequent activation of the nitrogen catabolite repression (NCR) response. Mislocalization of Gln3 occurs both in elp3∆ (removing the mcm5 moiety) and in urm∆ (s2 moiety). A link between t6A synthesis enzymes and TOR has also recently been seen in flies. The Glavic group showed that lowering levels of one of the subunits of the KEOPS complex (Bud32) reduces TOR phosphorylation of S6K (SCH9 in yeast) required for TOR dependent regulation of ribosome biogenesis [21].
–
How Gcn4 and TOR signaling depend on t6A and mcm5s2U is still far from understood at the molecular level. Are the Gcn4 activation and TOR repression in strains lacking these modifications due to direct effects caused by poor translation of specific proteins or are they part of general stress responses caused by translation inaccuracy and protein misfolding? The reality might lie in a combination of responses as in addition to the targeted effects described above, low mcm5s2U increases levels of proteins involved in proteasomal degradation [12]. In addition, s2 synthesis proteins in S. cerevisiae are unstable at high temperature and reduced levels of the modification lead to activation of the heat-shock response regulator (Hsf1) through the synthesis of unfolded proteins (Fig. 1) [22]. Finally, silencing both t6A synthesis genes Bud32 and Kae1 in flies activates the Unfolded Protein Response (UPR) [23] and mutations of the thiolation enzyme leading to the formation of ms2i6A in mouse led to an increase of the Endoplasmic Reticulum (ER) stress response [14].
–
Because the synthesis of the t6A and mcm5s2U modifications of tRNALysUUU draw on primary metabolism intermediates [24], it is tempting to propose that these could serve as sensing signals linking metabolism and translation. One recent example of such an integration is found in the mcm5s2U thiolation pathway; sulfur starvation reduces the levels of the Uba4 thiolation enzyme and hence the levels of mcm5s2U in yeast [13] (Fig. 1). Even if the underlying molecular mechanisms are not fully understood, low mcm5s2U levels trigger an adaptive response: 1) reduced protein expression due to general slow-down of translation of lysine rich proteins that are found predominantly in the ribosomal machinery; 2) increased levels of methionine, cysteine, and lysine synthesis proteins [13].
–
The complexity of the responses with the interplay of central regulators such as GCN4 and TOR (Fig. 1), make the dissection of the roles of t6A and mcm5s2U a delicate exercise that will require combining of classical genetic and biochemical studies with the genome wide bioinformatics, proteomic and profiling studies that are now available. These studies are all the more critical as derivatives of both modifications have been linked to human diseases as defects in the ms2t6A synthesis enzyme have been linked to type 2 diabetes [14], and familial dysautonomia patients have reduced levels of mcm5s2U [25].
References
- Z.D. Dalebroux, and M.S. Swanson, "ppGpp: magic beyond RNA polymerase", Nature Reviews Microbiology, vol. 10, pp. 203-212, 2012. http://dx.doi.org/10.1038/nrmicro2720
- N.J. Green, F.J. Grundy, and T.M. Henkin, "The T box mechanism: tRNA as a regulatory molecule", FEBS Letters, vol. 584, pp. 318-324, 2009. http://dx.doi.org/10.1016/j.febslet.2009.11.056
- A.G. Hinnebusch, "TRANSLATIONAL REGULATION OFGCN4AND THE GENERAL AMINO ACID CONTROL OF YEAST", Annual Review of Microbiology, vol. 59, pp. 407-450, 2005. http://dx.doi.org/10.1146/annurev.micro.59.031805.133833
- V. Scheidt, . , A. Juedes, C. Baer, R. Klassen, R. Schaffrath, and . , "Loss of wobble uridine modification in tRNA anticodons interferes with TOR pathway signaling", Microbial Cell, vol. 1, pp. 416-424, 2014. http://dx.doi.org/10.15698/mic2014.12.179
- B. El Yacoubi, M. Bailly, and V. de Crécy-Lagard, "Biosynthesis and Function of Posttranscriptional Modifications of Transfer RNAs", Annual Review of Genetics, vol. 46, pp. 69-95, 2012. http://dx.doi.org/10.1146/annurev-genet-110711-155641
- L. Perrochia, E. Crozat, A. Hecker, W. Zhang, J. Bareille, B. Collinet, H. van Tilbeurgh, P. Forterre, and T. Basta, "In vitro biosynthesis of a universal t6A tRNA modification in Archaea and Eukarya", Nucleic Acids Research, vol. 41, pp. 1953-1964, 2012. http://dx.doi.org/10.1093/nar/gks1287
- F. Meng, Y. Hu, N. Shen, X. Tong, J. Wang, J. Ding, and J. Zhou, "Sua5p a single-stranded telomeric DNA-binding protein facilitates telomere replication", The EMBO Journal, vol. 28, pp. 1466-1478, 2009. http://dx.doi.org/10.1038/emboj.2009.92
- C. Chen, B. Huang, M. Eliasson, P. Rydén, and A.S. Byström, "Elongator Complex Influences Telomeric Gene Silencing and DNA Damage Response by Its Role in Wobble Uridine tRNA Modification", PLoS Genetics, vol. 7, pp. e1002258, 2011. http://dx.doi.org/10.1371/journal.pgen.1002258
- A. Esberg, B. Huang, M.J. Johansson, and A.S. Byström, "Elevated Levels of Two tRNA Species Bypass the Requirement for Elongator Complex in Transcription and Exocytosis", Molecular Cell, vol. 24, pp. 139-148, 2006. http://dx.doi.org/10.1016/j.molcel.2006.07.031
- J. Fernández-Vázquez, I. Vargas-Pérez, M. Sansó, K. Buhne, M. Carmona, E. Paulo, D. Hermand, M. Rodríguez-Gabriel, J. Ayté, S. Leidel, and E. Hidalgo, "Modification of tRNALysUUU by Elongator Is Essential for Efficient Translation of Stress mRNAs", PLoS Genetics, vol. 9, pp. e1003647, 2013. http://dx.doi.org/10.1371/journal.pgen.1003647
- F. Bauer, A. Matsuyama, J. Candiracci, M. Dieu, J. Scheliga, D. Wolf, M. Yoshida, and D. Hermand, "Translational Control of Cell Division by Elongator", Cell Reports, vol. 1, pp. 424-433, 2012. http://dx.doi.org/10.1016/j.celrep.2012.04.001
- V.A.N. Rezgui, K. Tyagi, N. Ranjan, A.L. Konevega, J. Mittelstaet, M.V. Rodnina, M. Peter, and P.G.A. Pedrioli, "tRNA tK UUU , tQ UUG , and tE UUC wobble position modifications fine-tune protein translation by promoting ribosome A-site binding", Proceedings of the National Academy of Sciences, vol. 110, pp. 12289-12294, 2013. http://dx.doi.org/10.1073/pnas.1300781110
- S. Laxman, B. Sutter, X. Wu, S. Kumar, X. Guo, D. Trudgian, H. Mirzaei, and B. Tu, "Sulfur Amino Acids Regulate Translational Capacity and Metabolic Homeostasis through Modulation of tRNA Thiolation", Cell, vol. 154, pp. 416-429, 2013. http://dx.doi.org/10.1016/j.cell.2013.06.043
- F. Wei, T. Suzuki, S. Watanabe, S. Kimura, T. Kaitsuka, A. Fujimura, H. Matsui, M. Atta, H. Michiue, M. Fontecave, K. Yamagata, T. Suzuki, and K. Tomizawa, "Deficit of tRNALys modification by Cdkal1 causes the development of type 2 diabetes in mice", Journal of Clinical Investigation, vol. 121, pp. 3598-3608, 2011. http://dx.doi.org/10.1172/JCI58056
- M. Daugeron, T.L. Lenstra, M. Frizzarin, B. El Yacoubi, X. Liu, A. Baudin-Baillieu, P. Lijnzaad, L. Decourty, C. Saveanu, A. Jacquier, F.C.P. Holstege, V. de Crécy-Lagard, H. van Tilbeurgh, and D. Libri, "Gcn4 misregulation reveals a direct role for the evolutionary conserved EKC/KEOPS in the t6A modification of tRNAs", Nucleic Acids Research, vol. 39, pp. 6148-6160, 2011. http://dx.doi.org/10.1093/nar/gkr178
- B. Zinshteyn, and W.V. Gilbert, "Loss of a Conserved tRNA Anticodon Modification Perturbs Cellular Signaling", PLoS Genetics, vol. 9, pp. e1003675, 2013. http://dx.doi.org/10.1371/journal.pgen.1003675
- K. Natarajan, M.R. Meyer, B.M. Jackson, D. Slade, C. Roberts, A.G. Hinnebusch, and M.J. Marton, "Transcriptional Profiling Shows that Gcn4p Is a Master Regulator of Gene Expression during Amino Acid Starvation in Yeast", Molecular and Cellular Biology, vol. 21, pp. 4347-4368, 2001. http://dx.doi.org/10.1128/MCB.21.13.4347-4368.2001
- M.E. Cardenas, N.S. Cutler, M.C. Lorenz, C.J. Di Como, and J. Heitman, "The TOR signaling cascade regulates gene expression in response to nutrients", Genes & Development, vol. 13, pp. 3271-3279, 1999. http://dx.doi.org/10.1101/gad.13.24.3271
- D. Jablonowski, J. Täubert, C. Bär, M.J.R. Stark, and R. Schaffrath, "Distinct Subsets of Sit4 Holophosphatases Are Required for Inhibition ofSaccharomyces cerevisiaeGrowth by Rapamycin and Zymocin", Eukaryotic Cell, vol. 8, pp. 1637-1647, 2009. http://dx.doi.org/10.1128/EC.00205-09
- D. Jablonowski, L. Fichtner, M.J. Stark, and R. Schaffrath, "The Yeast Elongator Histone Acetylase Requires Sit4-dependent Dephosphorylation for Toxin-Target Capacity", Molecular Biology of the Cell, vol. 15, pp. 1459-1469, 2004. http://dx.doi.org/10.1091/mbc.E03-10-0750
- C. Ibar, V.F. Cataldo, C. Vásquez-Doorman, P. Olguín, and �. Glavic, "Drosophilap53-related protein kinase is required for PI3K/TOR pathway-dependent growth", Development, vol. 140, pp. 1282-1291, 2013. http://dx.doi.org/10.1242/dev.086918
- J.R. Damon, D. Pincus, and H.L. Ploegh, "tRNA thiolation links translation to stress responses inSaccharomyces cerevisiae", Molecular Biology of the Cell, vol. 26, pp. 270-282, 2015. http://dx.doi.org/10.1091/mbc.E14-06-1145
- D. Rojas-Benítez, C. Ibar, and �. Glavic, "TheDrosophilaEKC/KEOPS complex", Fly, vol. 7, pp. 168-172, 2013. http://dx.doi.org/10.4161/fly.25227
- M. Helm, and J. Alfonzo, "Posttranscriptional RNA Modifications: Playing Metabolic Games in a Cell’s Chemical Legoland", Chemistry & Biology, vol. 21, pp. 174-185, 2014. http://dx.doi.org/10.1016/j.chembiol.2013.10.015
- T. Karlsborn, H. Tükenmez, C. Chen, and A.S. Byström, "Familial dysautonomia (FD) patients have reduced levels of the modified wobble nucleoside mcm5s2U in tRNA", Biochemical and Biophysical Research Communications, vol. 454, pp. 441-445, 2014. http://dx.doi.org/10.1016/j.bbrc.2014.10.116
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (grant number R01 GM70641 to V.D.C.-L.), P.C.T. was funded in part by a Chateaubriand Fellowship from the French Embassy in the United States. We thank Jennifer Thiaville for editing the manuscript.
COPYRIGHT
© 2015

The emerging role of complex modifications of tRNALysUUU in signaling pathways by Patrick C. Thiaville and Valérie de Crécy-Lagard is licensed under a Creative Commons Attribution 4.0 International License.