Reviews:
Microbial Cell, Vol. 2, No. 9, pp. 308 - 321; doi: 10.15698/mic2015.09.224
The many facets of homologous recombination at telomeres
European Research Institute for the Biology of Ageing, University of Groningen, University Medical Center Groningen, Groningen, The Netherlands.
Keywords: homologous recombination, telomeres, alternative lengthening of telomeres, telomerase-independent telomere maintenance, break-induced replication.
Abbreviations:
ALT - alternative lengthening of telomeres,
BIR - break-induced replication,
DSB - DNA double-strand break,
HR- homologous recombination,
NHEJ - non-homologous end joining,
TERRA - telomeric repeat containing RNA,
TRD - telomere rapid deletion,
T-SCE - telomere sister chromatid exchange.
Received originally: 26/04/2015 Received in revised form: 26/06/2015
Accepted: 30/06/2015
Published: 30/07/2015
Correspondence:
Michael Chang, European Research Institute for the Biology of Ageing, University of Groningen, University Medical Center Groningen, Antonius Deusinglaan 1; Groningen 9713 AV, The Netherlands m.chang@umcg.nl
Conflict of interest statement: The authors declare that there are no conflicts of interest.
Please cite this article as: Clémence Claussin and Michael Chang (2015). The many facets of homologous recombination at telomeres. Microbial Cell 2(9): 308-321
Abstract
The ends of linear chromosomes are capped by nucleoprotein structures called telomeres. A dysfunctional telomere may resemble a DNA double-strand break (DSB), which is a severe form of DNA damage. The presence of one DSB is sufficient to drive cell cycle arrest and cell death. Therefore cells have evolved mechanisms to repair DSBs such as homologous recombination (HR). HR-mediated repair of telomeres can lead to genome instability, a hallmark of cancer cells, which is why such repair is normally inhibited. However, some HR-mediated processes are required for proper telomere function. The need for some recombination activities at telomeres but not others necessitates careful and complex regulation, defects in which can lead to catastrophic consequences. Furthermore, some cell types can maintain telomeres via telomerase-independent, recombination-mediated mechanisms. In humans, these mechanisms are called alternative lengthening of telomeres (ALT) and are used in a subset of human cancer cells. In this review, we summarize the different recombination activities occurring at telomeres and discuss how they are regulated. Much of the current knowledge is derived from work using yeast models, which is the focus of this review, but relevant studies in mammals are also included.
INTRODUCTION
Telomeres, nucleoprotein structures located at the ends of linear chromosomes, prevent natural chromosome ends from being recognized as DNA double-strand breaks (DSBs) (reviewed in [1]). Telomere dysfunction can lead to inappropriate repair activities, such as homologous recombination (HR) and non-homologous end joining (NHEJ). Such activities at telomeres can result in chromosomal rearrangements and genomic instability. Due to incomplete DNA replication and nucleolytic degradation, telomeres shorten with each round of replication, eventually leading to a growth arrest, known as replicative senescence, or to apoptosis. Telomere shortening can however be counteracted by a specialized reverse transcriptase called telomerase, which is composed of a protein catalytic subunit and an RNA subunit [2][3][4][5]. Telomerase extends telomeres by iterative reverse transcription of a short sequence to the 3‘ ends of telomeres, using the RNA subunit as a template [4][6][7].
–
Most human somatic cells do not express sufficient telomerase to prevent telomere shortening, which may be a contributing factor towards human ageing. This absence of telomere maintenance may have evolved as a barrier to tumorigenesis (reviewed in [8]). Indeed, cancer cells need to activate a telomere maintenance mechanism (TMM), and in approximately 85–90% of cancers this occurs through the upregulation of telomerase [9]. The remaining 10–15% of cancers employ telomerase-independent, recombination-based mechanisms, collectively termed alternative lengthening of telomeres (ALT) [10]. ALT mechanisms were first described as a TMM in the budding yeast Saccharomyces cerevisiae, where these cells are called “survivors” [11]. While recombination is clearly important for ALT and in survivors, recombination proteins are also important in non-ALT/survivor cells. For example, in S. cerevisiae, the combined absence of recombination and telomerase leads to a drastically enhanced rate of replicative senescence even though the rate of telomere shortening is apparently unchanged [11][12], although rare telomere loss events may be occurring. Furthermore, recombination proteins are important to resolve recombination intermediates at telomeres in pre-senescent cells [13], and can be detected at telomeres well before the appearance of survivors [14].
–
In this review, we will discuss how recombination is regulated at telomeres in telomerase-positive cells, telomerase-negative senescing cells, and telomerase-negative cells using recombination-mediated TMMs. Our focus will be on the significant advances made using different yeast models, but when appropriate, we will discuss relevant studies in mammalian systems.
SUPPRESSION OF HOMOLOGOUS RECOMBINATION AT TELOMERES
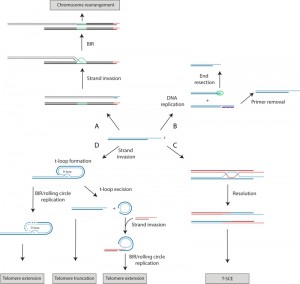
HR can be defined as the exchange of DNA sequences between two homologous DNA molecules and can be used to repair DNA damage, in particular DSBs. Although there are multiple variations regarding how HR can be used to repair a DSB, all of these models initiate with the resection of the 5‘ ends of the break to yield 3‘ single-stranded tails, of which one, or both, can invade homologous double-stranded DNA and prime DNA synthesis, templated by the donor double-stranded DNA (reviewed in [15]). These recombination intermediates are then processed by either helicases or resolvases, or both, to yield the final repaired product (Figure 1A). Ideally, both ends of a DSB remain in close proximity, but if this cannot be realized, a single end of a DSB can be repaired by an HR-mediated pathway termed break-induced replication (BIR). One-ended DSBs can also occur after the collapse of a replication fork. In BIR, the one-ended DSB invades a homologous sequence and replicates to the end of the invaded chromosome (Figure 1B). Since a BIR event could potentially result in extensive loss-of-heterozygosity, the BIR pathway is suppressed if both ends of a DSB are present [16]. Although a telomere resembles a resected one-ended DSB, there is no evidence that BIR is constitutively active in non-ALT/survivor cells, suggesting that BIR must also be suppressed at functional telomeres. This suppression may stem from a need to prevent telomeres from recombining with chromosome-internal telomeric sequences, as such events would lead to chromosomal rearrangements, and potentially to gene duplications (Figure 2A). Alternatively, the suppression of BIR may function to prevent inappropriate ALT/survivor-like telomere lengthening.
It is difficult to accurately measure telomere recombination events, due in part to the uniformity of telomeric repeats. Such events, however, can be detected in S. cerevisiae. This is possible because S. cerevisiae telomeres consist of imperfect, degenerate repeats [17][18], which is caused by telomerase only using a portion of the RNA template in each extension cycle, and because the RNA template and telomeric DNA can align in different registers [19]. Sequencing multiple copies of the same telomere derived from a clonal population of cells reveals a centromere-proximal region of stable sequence and a distal region with differing degenerate repeats [18][20]. This degenerate distal region is largely abolished in the absence of telomerase [20], but rare sequence divergence events can be detected [21]. Presumably, such events are occurring in the presence of telomerase as well, but it is possible that telomerase can influence recombination activity. These telomerase-independent events are thought to be due to unequal sister chromatid exchange or intertelomeric recombination, and occur at a frequency of less than 0.3% per telomere per generation [21]. We have recently conducted more careful measurements indicating that the frequency of these events may even be substantially lower than 0.3% (C. Claussin and M. Chang, unpublished data), suggesting that while recombination proteins are important during senescence, as mentioned above, their activity does not result in unequal sister chromatid exchange and intertelomeric recombination events, as measured in this assay. Thus these events are normally tightly repressed until the emergence of survivors, when such events can readily be detected [22].
The mechanism by which recombination of telomeric repeats is suppressed in S. cerevisiae is not entirely clear. Proteins that are present at a telomere but not at a DSB make obvious candidates to mediate the suppression (Figure 3A). The double-stranded portion of the telomere is bound by Rap1 [23], which recruits the additional factors Rif1 and Rif2 [24][25], as well as the silent chromatin proteins, Sir3 and Sir4 [26]. The CST complex (consisting of Cdc13, Stn1 and Ten1) binds to the 3‘ overhang [27][28][29][30][31].
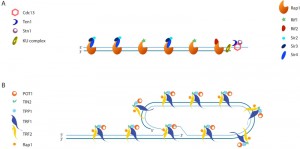 | FIGURE 3. (A) Schematic of an S. cerevisiae telomere with associated proteins. (B) Schematic of shelterin-bound mammalian telomeres shown in a t-loop configuration. |
Using the telomere sequencing approach described above, one study reported that the deletion of RIF1 may increase telomeric recombination events, particularly at telomeres less than 120 bp in length [21], but a subsequent study failed to confirm this finding [22]. Neither study found any role of Rif2 in suppressing recombination. A separate genetic assay designed to detect telomeric recombination events also found no role of Rif1, Rif2, or the Sir proteins in these events [32]. In contrast, cdc13-1 and stn1-13 mutant strains exhibit elevated levels of telomeric recombination [30][32][33]. The Ku heterodimer (consisting of Yku70 and Yku80) has also been shown to inhibit recombination at telomeres [32][34]. Ku functions at both DSBs and telomeres to inhibit 5’–3’ end resection, and accordingly, cells lacking Ku have increased 3‘ telomeric overhangs [34][35][36][37]. The cdc13-1 and stn1-13 mutations also result in extensive telomere resection and long 3’ overhangs [29][33]. Since end resection is the first step in the processing of a DSB for subsequent recombination, these findings suggest that the CST and Ku complexes suppress telomeric recombination by inhibiting end resection at telomeres. As cdc13-1 yku∆ double mutants exhibit synthetic growth defects [34][38] and senesce after ~25 generations [32], it is likely that Cdc13 and the Ku complex function in separate pathways to inhibit resection. Consistent with this idea, Ku is more important for inhibiting resection in G1 and CST is more important in the S/G2 phases of the cell cycle [39]
–
While excessive telomeric resection is detrimental, some resection is needed to generate a 3’ overhang (Figure 2B), which is needed for proper telomere capping. The 3’ overhang is also the substrate for telomerase activity [40]. In S. cerevisiae, end resection is initiated by the conserved MRX/N complex (Mre11, Rad50, and Xrs2 in S. cerevisiae, Nbs1 in other organisms), which together with Sae2 (CtIP in other organisms) can perform limited end resection [41][42]. Following this initial step, more extensive resection is carried out by the 5’–3’ exonuclease, Exo1, or the combined activities of the Sgs1-Top3-Rmi1 complex and Dna2 [41][42] (Figure 1A). The Ku and CST complexes inhibit this more extensive resection. Rap1 and its associated protein Rif2 are also important for limiting MRX-mediated telomeric resection [37].
–
Similar to S. cerevisiae, telomeric recombination can be induced by perturbing telomere capping proteins in Kluyveromyces lactis. A mutation in the gene encoding K. lactis Stn1 (stn1-M1) causes survivor-like recombination-mediated telomere elongation, even in the presence of functional telomerase [43]. Furthermore, telomerase-negative K. lactis mutants containing mutant repeats that disrupt Rap1 binding develop even longer telomeres than if they would have wild-type repeats, suggesting that Rap1 also plays a role in repressing recombination [44].
–
In addition to recombination of the telomeric tracts, the Ku complex also suppresses subtelomeric recombination in both S. cerevisiae [45] and Schizosaccharomyces pombe [46], although it is unclear how similar the mechanisms governing telomeric and subtelomeric recombination are. S. pombe cells lacking Taz1 (ortholog of mammalian shelterin components, TRF1 and TRF2; see below), which binds to the double-stranded portion of fission yeast telomeres, also exhibit elevated levels of subtelomeric recombination, which is thought to be a response to increased replication fork stalling [47].
–
In mammals, telomeric recombination, as measured by telomere sister chromatid exchange events (T-SCEs; Figure 2C), is inhibited by the combined action of POT1, TRF2, RAP1, and KU. Like Cdc13, POT1 binds to single-stranded telomeric DNA. Unlike yeast Rap1, human RAP1 has relatively weak DNA binding activity [48] and is instead recruited to telomeres by TRF2, which binds to double-stranded telomeric DNA. POT1, TRF2, and RAP1, along with TRF1, TIN2, and TPP1, form a six-membered telomere-capping complex called shelterin (Figure 3B; mouse shelterin has two POT1 orthologs (POT1a and POT1b) due to a recent duplication within the rodent lineage; reviewed in [1]). Although loss of TRF2 or KU alone, or loss of both POT1a and POT1b together, exhibit only basal levels of T-SCEs (~1.5–3% of telomeres) in mouse embryonic fibroblasts (MEFs), T-SCEs are seen at approximately 10–15% of telomeres in MEFs lacking both TRF2 and KU, or in triple knockouts lacking POT1a, POT1b, and KU [49][50]. The role of TRF2 in suppressing T-SCEs may be mediated by its recruitment of RAP1, as KU-deficient MEFs expressing an allele of TRF2 that cannot bind to RAP1 also exhibit elevated T-SCE levels [51].
SOME RECOMBINATIONAL ACTIVITIES AT TELOMERES ARE BENEFICIAL
Although full activation of HR pathways must be prevented at telomeres, some recombination processes appear to be required for proper telomere function. In mammals, the 3’ telomeric overhang can be further protected within a t-loop configuration (Figure 2D and Figure 3). A t-loop is a lariat structure formed by the invasion of the 3’ overhang into the double-stranded portion of the same telomere. TRF2 is required for the formation and/or maintenance of t-loops [52][53]. HR factors may be needed for the strand invasion step, as RAD51 and its paralog XRCC3, along with RAD52, can be detected at telomeres after replication, and these proteins are required for the generation of telomeric D-loops in an in vitro assay [54]. However, recent biochemical studies indicate that TRF2 actually inhibits RAD51-mediated D-loop formation [55], and it also recruits the helicase RTEL1 to promote t-loop unwinding in S phase [56], indicating that TRF2 is both a positive and a negative regulator of t-loops. It has been proposed that the t-loop is important for disguising the chromosome ends, preventing the activation of the ATM checkpoint kinase and NHEJ [57], although it is possible that TRF2 directly inhibits ATM independently of t-loop formation [58]. However, a t-loop also resembles an HR intermediate, which could lead to the formation of a Holliday junction, and thus has the potential to be excised through the action of resolvases, resulting in rapid telomere shortening (Figure 2D). Such excisions can occur in TRF2 mutants lacking its N-terminal basic domain (TRF2∆B) through a process that requires XRCC3 [59]. XRCC3 and RAD51C, both of whom are RAD51 paralogs, form a complex that is associated with Holliday junction resolvase activity in vitro [60]. Furthermore, the N-terminal basic domain was recently found to inhibit GEN1 and MUS81, two endonucleases with resolvase activity [61]. Thus, TRF2 is important for both the formation of t-loops and for preventing their excision from telomeres.
–
Unlike mammalian t-loops, yeast t-loops have been difficult to observe by electron microscopy because yeast telomeres are short, and long telomere restriction fragments, which can be separated from short genomic restriction fragments, are required for the visualization of native telomeric DNA. However, t-loops have been observed in K. lactis strains with elongated telomeres [62], and extrachromosomal DNA circles containing telomeric repeats, hypothesized to be excised t-loops, can be found in certain cell types, such as S. cerevisiae survivors [63][64].
–
While t-loop excision is normally repressed, over-elongated telomeres are specifically targeted for shortening by a mechanism called telomere rapid deletion (TRD), which is thought to occur through the excision of t-loops [65]. TRD, also referred to as telomere trimming [66], was first seen in S. cerevisiae, where it was demonstrated that an over-elongated telomere can be shortened to wild-type length via a t-loop-excising intrachromosomal recombination event [67][68]. TRD has been observed in K. lactis [69], Arabidopsis thalania [70], and human cells [66][71]. Whether over-elongated telomeres are actively targeted for shortening by TRD is unclear, and the mechanism by which short or wild-type length telomeres, but not over-elongated telomeres, are protected from TRD has yet to be elucidated.
–
Proteins involved in HR are also required for proper replication of telomeres. Recombination processes are important for dealing with stalled or collapsed replication forks (reviewed in [72]), and telomere sequences are known to cause problems for the replication machinery in S. cerevisiae [73], S. pombe [74], and mammals [75]. Mouse cells lacking BRCA2, RAD51, RAD51C, RAD51D, and RAD54 have short telomeres and show signs of telomere fragility [76][77][78].
–
One reason that telomeres are difficult to replicate may be due to the transcription of telomeres, which produces long non-coding RNA called TERRA (telomeric repeat containing RNA). Co-transcriptionally-formed RNA-DNA hybrids (also referred to as R-loops) or the RNA polymerase II machinery itself can hinder DNA replication fork progression, which can lead to transcription-associated recombination (reviewed in [79]). In S. cerevisiae, increasing the rate of telomere transcription induces Exo1-mediated telomere resection, which promotes telomeric recombination [80]. Likewise, TERRA RNA-DNA hybrids, which can be resolved by RNase H and the THO complex (named after one of its subunits, Tho2), can also induce recombination. Mutating either RNase H or the THO complex increases the abundance of RNA-DNA hybrids at telomeres, leading to an increase in telomeric recombination [81][82][83]. Thus, both the process of transcribing telomeres and TERRA R-loops can independently lead to telomeric recombination, which may be needed to preserve telomere integrity, especially in the absence of telomerase [80].
TELOMERE MAINTENANCE VIA RECOMBINATION
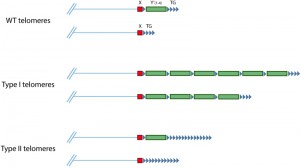
In the absence of telomerase, telomeres shorten until they activate the DNA damage checkpoint, which in turn stops further cell proliferation. However, some cells can overcome this barrier by using recombination-mediated mechanisms to elongate their telomeres. Such cells were first discovered in S. cerevisiae, where they are called survivors [11]. There are two main types of survivors: type I survivors exhibit amplification of Y’ subtelomeric elements, while type II survivors exhibit amplification of the terminal telomeric repeats (Figure 4) [11][84]. Both types require Rad52, needed for almost all recombination in S. cerevisiae, and Pol32, a non-essential subunit of DNA polymerase d that is required for BIR [11][85]. The importance of Pol32 indicates that, in the absence of telomerase, BIR-mediated mechanisms can maintain telomeres, and that the suppression of BIR at telomeres must be alleviated in survivors. Deletion of PIF1 also greatly impairs the formation of both types of survivors [86][87], which is likely attributable to the role of Pif1 in BIR [88]. BIR can take place in a Rad51-dependent manner, or in a Rad51-independent manner that requires the MRX complex and Rad59 [89][90]. In addition to Rad52 and Pol32, type I survivors require Rad51, Rad54, and Rad57, whereas type II survivors require the MRX complex and Rad59 instead [12][91]. This strongly suggests that telomere maintenance in type I survivors involve Rad51-dependent BIR while Rad51-independent BIR is important for type II survivors.
Sgs1 and Exo1 are also important for type II survivor formation [92][93][94][95]. Sgs1 and Exo1 are needed for processive resection of DNA ends [41][42], suggesting that end resection might promote type II survivor formation by helping to generate a 3’ overhang to initiate BIR. Consistent with this idea, an sgs1-D664∆ mutation, which is still competent in recombination repair but defective in resection [96][97], also prevents type II survivor formation [98]. Interestingly, extensive resection by Sgs1 and Exo1 inhibits Rad51-dependent BIR [99][100]. Thus, deletion of SGS1 or EXO1 may both promote the Rad51-dependent type I pathway and disrupt the Rad51-independent type II pathway. Similarly, deletion of FUN30, which encodes a chromatin remodeler that promotes end resection, partially hinders the formation of type II survivors [101].
–
A number of additional proteins have been implicated in the generation of type II survivors. These include the DNA damage checkpoint kinases Mec1 and Tel1 [102], the B-type cyclin Clb2 [103], Def2, an RNA polymerase II degradation factor, [104], Mdt4/Pin4, a protein that interacts with the checkpoint kinase Rad53 [105], and Sua5, a protein required for an essential tRNA modification [106]. A recent screen identified a further 22 genes important for type II survivor formation, including genes encoding for members of the KEOPS complex, the Rad6 DNA repair pathway, and proteins involved in nonsense-mediated decay [87]. The same screen also identified that the INO80 chromatin remodeling complex affects the emergence of type I survivors [87]. Exactly how these genes affect survivor formation is unclear.
–
Telomere length just before the emergence of survivors has also been shown to affect the ratio of type I to type II survivors formed, with long telomeres favoring type II survivors [22]. Deletion of RIF1 and RIF2 strongly biases toward type II survivor formation [107], which is likely due to telomerase-negative rif mutants senescing with longer telomeres than telomerase-negative RIF strains [22]. However, it is also possible that the Rif proteins limit type II survivor formation by inhibiting end resection [37].
–
Type I survivors typically arise more frequently but grow very poorly, whereas the growth of type II survivors is comparable to telomerase-positive cells [11][84]. Rad51-dependent BIR is more efficient than Rad51-independent BIR [16][89], which may explain the higher frequency of type I survivors. The poor growth of type I survivors may be due to the maintenance of very short telomeres in these cells. Like senescent cells, type I survivors are arrested at the G2/M boundary with telomeres moving back and forth between the mother and the bud [108], indicating that the telomeres of type I survivors do not return to a properly capped state. In contrast, the long, heterogeneously sized telomeres of type II survivors behave in this respect like telomeres in telomerase-positive cells [108].
–
In type I survivors, all telomeres are extended through the amplification of subtelomeric Y’ elements, even telomeres that did not originally have Y’ elements [11]. The movement of Y’ sequences among chromosome ends can be explained as a BIR event initiated by an uncapped telomere that invades TG1-3 repeats that are found between some tandem Y’ elements, or between some X and Y’ elements (Figure 4) [109]. While the formation of type I survivors is Rad51-dependent, efficient movement of Y’ elements in the senescing phase before the emergence of survivors is facilitated more so by Rad59 than Rad51 [110], which is surprising given that Rad59 is not required for type I survivor formation. The Y’ element encodes a poorly characterized helicase that is strongly induced in type I survivors and this helicase may be important for the viability of type I survivors [111]. Thus, amplification of the Y’ elements may facilitate this process, but it is also possible that Y’ amplification is only needed to provide homologous sequences at every telomere to allow for more efficient BIR. It is also unclear how the short terminal TG1-3 sequences are maintained.
–
Type II survivors are thought to elongate telomeres through a ‘roll-and-spread’ mechanism, involving both rolling circle synthesis and intertelomeric BIR events [112]. Support for such a model is largely based on studies from K. lactis, where all survivors are type II due to a lack of subtelomeric blocks of telomeric repeats to allow for a type I-like pathway [113]. K. lactis survivors derived from cells with two kinds of telomere repeats (i.e. repeats that are either wild type or mutant in sequence) usually contain repeating patterns in the lengthened telomeres, most likely arising from small circles containing telomere DNA being used as templates for rolling circle replication [114]. Furthermore, transformation of a DNA circle containing mutant telomere repeats into a K. lactis telomerase-negative strain results in the incorporation of long tandem arrays of the mutant repeats at telomeres [114]. These observations led to a model whereby a circle containing telomeric DNA (i.e. a t-circle) is produced by a recombination event, possibly through the excision of a t-loop [62]. An uncapped telomere can then initiate BIR-mediated rolling circle DNA synthesis using the t-circle as a template (Figure 2D). In addition to being observed in K. lactis [115], t-circles are also found in S. cerevisiae survivors [63][64] and human ALT cells [59][116].
–
S. pombe can also form telomerase-negative survivors either by circularizing their three chromosomes (in a process that involves either NHEJ or the single-strand annealing recombination pathway; reviewed in [117]) or, much less frequently, by maintenance of linear chromosomes through telomere recombination [118]. Deletion of taz1+ greatly increases survival with linear chromosomes, which allows easier examination of this pathway, and suggests that Taz1 inhibits recombination at telomeres [118]. Rad22 (S. pombe Rad52), Tel1, and the MRN complex are required for telomere maintenance in telomerase-negative taz1∆ “linear” survivors, suggesting that these cells are similar to S. cerevisiae type II survivors [119].
–
Although Rad52 is a critical protein in the formation of survivors, rare Rad52-independent survivors can arise at a very low frequency in K. lactis [113], and in S. cerevisiae with long telomeres [120][121]. Like type II survivors, these Rad52-independent survivors rely on the amplification of the telomeric repeats. Long telomeres are preferentially elongated in emerging type II survivors [22], so longer telomeres may provide better substrates for recombination, allowing for recombination to happen even in the absence of Rad52. Consistent with this idea, single-strand annealing becomes Rad52-independent when homologous regions are several kilobases long [122], indicating that larger regions of homology can compensate for the lack of Rad52. Another class of Rad52-independent survivors can also occur in cells lacking Exo1 or Sgs1, and thus defective in end-resection [123][124]. These survivors have lost telomeric and subtelomeric sequences, but survive by forming large palindromes at chromosome ends.
RECOMBINATION-MEDIATED TELOMERE MAINTENANCE IN HUMAN CELLS
Although a type I-like ALT cell line has been reported [125][126], most human ALT cancer cells are thought to maintain their telomeres using recombination-mediated mechanisms that resemble what occurs in yeast type II survivors. Much like type II survivors, ALT cells often have long, heterogeneous-sized telomeres [10][127], abundant extrachromosomal DNA circles containing telomeric repeats [59][116][128], and a requirement for the MRN complex and the Sgs1-homolog, BLM, for telomere maintenance [129][130][131]. WRN, another Sgs1 homolog, is required for telomere maintenance in some, but not all, ALT cell lines, suggesting the existence of different ALT mechanisms [132]. Many ALT cells also possess special promyelocytic leukemia (PML) bodies, termed ALT-associated PML bodies [133] that contain telomeric DNA, shelterin proteins, and DNA damage response and HR factors, including RAD51, RAD52, BLM, WRN, and the MRN complex (reviewed in [134] and [135]). Furthermore, ALT cells exhibit an elevated frequency of T-SCEs [136][137].
–
Interestingly, although several shelterin components have been shown to inhibit telomeric recombination in telomerase-positive mammalian cells (discussed above), the abundance of all six shelterin proteins were unchanged in 22 different ALT cell lines, and exome sequencing failed to detect mutations in any of the genes encoding shelterin proteins [138]. The same study found the loss of chromatin-remodeler ATRX in 19 of the 22 cell lines. However, downregulation of ATRX is not sufficient to activate ALT, suggesting that loss of ATRX may only be one step in the process [138]. A recent study has found that ATRX represses telomere-bound TERRA in G2/M [139]. TERRA, in turn, inhibits hnRNPA1-mediated removal of RPA from telomeres in early S phase, but this inhibition is alleviated in late S phase when TERRA declines at telomeres [140]. Thus, ATRX deficiency leads to persistent association of TERRA, and consequently RPA, with telomeres, creating a recombinogenic structure that favors ALT [139]. Consistent with this model, TERRA levels are upregulated in ALT cells [138][141]. Increased TERRA transcription in S. cerevisiae is also thought to promote survivor formation [80], and RNA-DNA hybrids have also been demonstrated to promote the emergence of type II survivors [83].
–
Another recent study has found that co-depletion of the histone chaperone paralogs ASF1a and ASF1b induces most of the characteristics of ALT cells, including formation of ALT-associated promyelocytic leukemia bodies, presence of extrachromosomal telomeric DNA, increased T-SCEs, and greater telomere length heterogeneity [142]. One commonality of ATRX and ASF1 is that they both act on the histone variant H3.3. ATRX, together with DAXX, act together to deposit H3.3 at specific heterochromatic loci, such as telomeres, in a replication-independent manner [143][144][145][146][147]. Loss of ATRX or DAXX would impair H3.3 loading at telomeres, and mutations in the genes encoding ATRX, DAXX, and H3.3 are associated with ALT cancers [138][148][149][150]. ASF1, originally identified in S. cerevisiae [151], is a histone chaperone involved in both the replication-coupled and replication-independent incorporation of H3.1-H4 and H3.3-H4 histone dimers into nucleosomes [152][153]. It remains to be seen if and how ATRX and ASF1 act together to regulate recombination at telomeres.
TELOMERE LENGTH AND RECOMBINATION
Short telomeres are more likely to become dysfunctional and may therefore be more susceptible to HR activities. Indeed, analysis of telomere recombination events in telomerase-negative senescing S. cerevisiae cells using the telomere sequencing approach described above has revealed a preference for the recombination of short telomeres [21][22][154], and RNA-DNA hybrids formed by TERRA stimulates recombination at short telomeres [81]. Short telomeres are also preferentially elongated by recombination in established type II survivors [107][154]. In K. lactis, short telomeres increase subtelomeric recombination [155], and in senescing cells with a single long telomere, that long telomere almost always serves as the template for BIR initiated by the other shorter telomeres [156].
–
In contrast, long telomeres are preferentially extended in emerging type II S. cerevisiae survivors [22]. This finding is consistent with previous studies in prokaryotes, yeast, and mammalian cells showing that the efficiency of HR is proportional to the length of the substrate DNA [157][158][159][160][161][162]. The difference in the length preference may be due to which telomeres are in a recombination-competent state. Increasing telomere length may increase the likelihood of recombination, but only if the telomere in question is recombination-competent. In most situations, including in pre-senescent cells and in established survivors, only one or a few short telomeres are in an uncapped state, susceptible to recombination. However, senescent cells primed to become survivors likely have most or all of their telomeres sufficiently eroded and in a recombination-competent state.
CONCLUDING REMARKS
In this review, we have highlighted the many ways in which HR activities are important at telomeres. However, it is likely that there are additional facets of telomeric HR that have not been explored yet, and much work still needs to be done to determine how these activities are regulated. For example, it is still unclear how some HR activities are suppressed at telomeres (e.g. BIR, extensive end-resection, excision of t-loops, etc.) while others are not (e.g. initial end-resection, t-loop formation, TRD of over-elongated telomeres, etc.). Furthermore, how is the suppression of BIR alleviated in survivor/ALT cells? Considering the importance of HR pathways and telomere biology with respect to cancer and ageing, a better understanding of HR activities at telomeres will have broad ramifications for human health.
References
- D. Jain, and J.P. Cooper, "Telomeric Strategies: Means to an End", Annual Review of Genetics, vol. 44, pp. 243-269, 2010. http://dx.doi.org/10.1146/annurev-genet-102108-134841
- C.W. Greider, and E.H. Blackburn, "Identification of a specific telomere terminal transferase activity in tetrahymena extracts", Cell, vol. 43, pp. 405-413, 1985. http://dx.doi.org/10.1016/0092-8674(85)90170-9
- C.W. Greider, and E.H. Blackburn, "The telomere terminal transferase of tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity", Cell, vol. 51, pp. 887-898, 1987. http://dx.doi.org/10.1016/0092-8674(87)90576-9
- C.W. Greider, and E.H. Blackburn, "A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis", Nature, vol. 337, pp. 331-337, 1989. http://dx.doi.org/10.1038/337331a0
- J. Lingner, T.R. Hughes, A. Shevchenko, M. Mann, V. Lundblad, and T.R. Cech, "Reverse Transcriptase Motifs in the Catalytic Subunit of Telomerase", Science, vol. 276, pp. 561-567, 1997. http://dx.doi.org/10.1126/science.276.5312.561
- G. Yu, J.D. Bradley, L.D. Attardi, and E.H. Blackburn, "In vivo alteration of telomere sequences and senescence caused by mutated Tetrahymena telomerase RNAs", Nature, vol. 344, pp. 126-132, 1990. http://dx.doi.org/10.1038/344126a0
- M.S. Singer, and D.E. Gottschling, "TLC1 : Template RNA Component of Saccharomyces cerevisiae Telomerase", Science, vol. 266, pp. 404-409, 1994. http://dx.doi.org/10.1126/science.7545955
- S.E. Artandi, and R.A. DePinho, "Telomeres and telomerase in cancer", Carcinogenesis, vol. 31, pp. 9-18, 2009. http://dx.doi.org/10.1093/carcin/bgp268
- J. Shay, and S. Bacchetti, "A survey of telomerase activity in human cancer", European Journal of Cancer, vol. 33, pp. 787-791, 1997. http://dx.doi.org/10.1016/S0959-8049(97)00062-2
- T.M. Bryan, A. Englezou, L. Dalla-Pozza, M.A. Dunham, and R.R. Reddel, "Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines", Nature Medicine, vol. 3, pp. 1271-1274, 1997. http://dx.doi.org/10.1038/nm1197-1271
- V. Lundblad, and E.H. Blackburn, "An alternative pathway for yeast telomere maintenance rescues est1− senescence", Cell, vol. 73, pp. 347-360, 1993. http://dx.doi.org/10.1016/0092-8674(93)90234-H
- S. Le, J.K. Moore, J.E. Haber, and C.W. Greider, "RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase.", Genetics, 1999. http://www.ncbi.nlm.nih.gov/pubmed/10224249
- J.Y. Lee, M. Kozak, J.D. Martin, E. Pennock, and F.B. Johnson, "Evidence That a RecQ Helicase Slows Senescence by Resolving Recombining Telomeres", PLoS Biology, vol. 5, pp. e160, 2007. http://dx.doi.org/10.1371/journal.pbio.0050160
- B. Khadaroo, M.T. Teixeira, P. Luciano, N. Eckert-Boulet, S.M. Germann, M.N. Simon, I. Gallina, P. Abdallah, E. Gilson, V. Géli, and M. Lisby, "The DNA damage response at eroded telomeres and tethering to the nuclear pore complex", Nature Cell Biology, vol. 11, pp. 980-987, 2009. http://dx.doi.org/10.1038/ncb1910
- L.S. Symington, R. Rothstein, and M. Lisby, "Mechanisms and Regulation of Mitotic Recombination in Saccharomyces cerevisiae", Genetics, vol. 198, pp. 795-835, 2014. http://dx.doi.org/10.1534/genetics.114.166140
- A. Malkova, M.L. Naylor, M. Yamaguchi, G. Ira, and J.E. Haber, "RAD51-Dependent Break-Induced Replication Differs in Kinetics and Checkpoint Responses from RAD51-Mediated Gene Conversion", Molecular and Cellular Biology, vol. 25, pp. 933-944, 2005. http://dx.doi.org/10.1128/MCB.25.3.933-944.2005
- J. Shampay, J.W. Szostak, and E.H. Blackburn, "DNA sequences of telomeres maintained in yeast", Nature, vol. 310, pp. 154-157, 1984. http://dx.doi.org/10.1038/310154a0
- S.S. Wang, and V.A. Zakian, "Sequencing of Saccharomyces telomeres cloned using T4 DNA polymerase reveals two domains.", Molecular and cellular biology, 1990. http://www.ncbi.nlm.nih.gov/pubmed/2196453
- K. Förstemann, and J. Lingner, "Molecular Basis for Telomere Repeat Divergence in Budding Yeast", Molecular and Cellular Biology, vol. 21, pp. 7277-7286, 2001. http://dx.doi.org/10.1128/MCB.21.21.7277-7286.2001
- K. Forstemann, "Telomerase-dependent repeat divergence at the 3' ends of yeast telomeres", Nucleic Acids Research, vol. 28, pp. 2690-2694, 2000. http://dx.doi.org/10.1093/nar/28.14.2690
- M. Teixeira, M. Arneric, P. Sperisen, and J. Lingner, "Telomere Length Homeostasis Is Achieved via a Switch between Telomerase- Extendible and -Nonextendible States", Cell, vol. 117, pp. 323-335, 2004. http://dx.doi.org/10.1016/S0092-8674(04)00334-4
- M. Chang, J.C. Dittmar, and R. Rothstein, "Long telomeres are preferentially extended during recombination-mediated telomere maintenance", Nature Structural & Molecular Biology, vol. 18, pp. 451-456, 2011. http://dx.doi.org/10.1038/nsmb.2034
- E. Gilson, M. Roberge, R. Giraldo, D. Rhodes, and S.M. Gasser, "Distortion of the DNA Double Helix by RAP1 at Silencers and Multiple Telomeric Binding Sites", Journal of Molecular Biology, vol. 231, pp. 293-310, 1993. http://dx.doi.org/10.1006/jmbi.1993.1283
- C.F. Hardy, L. Sussel, and D. Shore, "A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation.", Genes & Development, vol. 6, pp. 801-814, 1992. http://dx.doi.org/10.1101/gad.6.5.801
- D. Wotton, and D. Shore, "A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae.", Genes & Development, vol. 11, pp. 748-760, 1997. http://dx.doi.org/10.1101/gad.11.6.748
- P. Moretti, K. Freeman, L. Coodly, and D. Shore, "Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1.", Genes & Development, vol. 8, pp. 2257-2269, 1994. http://dx.doi.org/10.1101/gad.8.19.2257
- J. Lin, and V. Zakian, "The Saccharomyces CDC 13 protein is a single-strand TG 1–3 telomeric DNA-binding protein in vitro that affects telomere behavior in vivo", Proceedings of the National Academy of Sciences, vol. 93, pp. 13760-13765, 1996. http://dx.doi.org/10.1073/pnas.93.24.13760
- C.I. Nugent, T.R. Hughes, N.F. Lue, and V. Lundblad, "Cdc13p: A Single-Strand Telomeric DNA-Binding Protein with a Dual Role in Yeast Telomere Maintenance", Science, vol. 274, pp. 249-252, 1996. http://dx.doi.org/10.1126/science.274.5285.249
- N. Grandin, S.I. Reed, and M. Charbonneau, "Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13.", Genes & Development, vol. 11, pp. 512-527, 1997. http://dx.doi.org/10.1101/gad.11.4.512
- N. Grandin, "Cdc13 prevents telomere uncapping and Rad50-dependent homologous recombination", The EMBO Journal, vol. 20, pp. 6127-6139, 2001. http://dx.doi.org/10.1093/emboj/20.21.6127
- H. Gao, R.B. Cervantes, E.K. Mandell, J.H. Otero, and V. Lundblad, "RPA-like proteins mediate yeast telomere function", Nature Structural & Molecular Biology, vol. 14, pp. 208-214, 2007. http://dx.doi.org/10.1038/nsmb1205
- M.L. DuBois, Z.W. Haimberger, M.W. McIntosh, and D.E. Gottschling, "A quantitative assay for telomere protection in Saccharomyces cerevisiae.", Genetics, 2002. http://www.ncbi.nlm.nih.gov/pubmed/12136006
- B. Garvik, M. Carson, and L. Hartwell, "Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint.", Molecular and cellular biology, 1995. http://www.ncbi.nlm.nih.gov/pubmed/7565765
- R.M. Polotnianka, J. Li, and A.J. Lustig, "The yeast Ku heterodimer is essential for protection of the telomere against nucleolytic and recombinational activities", Current Biology, vol. 8, pp. 831-835, 1998. http://dx.doi.org/10.1016/s0960-9822(98)70325-2
- S. Gravel, M. Larrivée, P. Labrecque, and R.J. Wellinger, "Yeast Ku as a Regulator of Chromosomal DNA End Structure", Science, vol. 280, pp. 741-744, 1998. http://dx.doi.org/10.1126/science.280.5364.741
- E.P. Mimitou, and L.S. Symington, "Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2", The EMBO Journal, vol. 29, pp. 3358-3369, 2010. http://dx.doi.org/10.1038/emboj.2010.193
- D. Bonetti, M. Clerici, S. Anbalagan, M. Martina, G. Lucchini, and M.P. Longhese, "Shelterin-Like Proteins and Yku Inhibit Nucleolytic Processing of Saccharomyces cerevisiae Telomeres", PLoS Genetics, vol. 6, pp. e1000966, 2010. http://dx.doi.org/10.1371/journal.pgen.1000966
- C.I. Nugent, G. Bosco, L.O. Ross, S.K. Evans, A.P. Salinger, J. Moore, J.E. Haber, and V. Lundblad, "Telomere maintenance is dependent on activities required for end repair of double-strand breaks", Current Biology, vol. 8, pp. 657-662, 1998. http://dx.doi.org/10.1016/s0960-9822(98)70253-2
- M.D. Vodenicharov, N. Laterreur, and R.J. Wellinger, "Telomere capping in non-dividing yeast cells requires Yku and Rap1", The EMBO Journal, vol. 29, pp. 3007-3019, 2010. http://dx.doi.org/10.1038/emboj.2010.155
- J. Lingner, and T.R. Cech, "Purification of telomerase from Euplotes aediculatus: requirement of a primer 3' overhang.", Proceedings of the National Academy of Sciences, vol. 93, pp. 10712-10717, 1996. http://dx.doi.org/10.1073/pnas.93.20.10712
- E.P. Mimitou, and L.S. Symington, "Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing", Nature, vol. 455, pp. 770-774, 2008. http://dx.doi.org/10.1038/nature07312
- Z. Zhu, W. Chung, E.Y. Shim, S.E. Lee, and G. Ira, "Sgs1 Helicase and Two Nucleases Dna2 and Exo1 Resect DNA Double-Strand Break Ends", Cell, vol. 134, pp. 981-994, 2008. http://dx.doi.org/10.1016/j.cell.2008.08.037
- S. Iyer, A.D. Chadha, and M.J. McEachern, "A Mutation in the STN1 Gene Triggers an Alternative Lengthening of Telomere-Like Runaway Recombinational Telomere Elongation and Rapid Deletion in Yeast", Molecular and Cellular Biology, vol. 25, pp. 8064-8073, 2005. http://dx.doi.org/10.1128/MCB.25.18.8064-8073.2005
- L.H. Bechard, B.D. Butuner, G.J. Peterson, W. McRae, Z. Topcu, and M.J. McEachern, "Mutant Telomeric Repeats in Yeast Can Disrupt the Negative Regulation of Recombination-Mediated Telomere Maintenance and Create an Alternative Lengthening of Telomeres-Like Phenotype", Molecular and Cellular Biology, vol. 29, pp. 626-639, 2009. http://dx.doi.org/10.1128/MCB.00423-08
- M.E. Marvin, C.D. Griffin, D.E. Eyre, D.B.H. Barton, and E.J. Louis, "InSaccharomyces cerevisiae, yKu and Subtelomeric Core X Sequences Repress Homologous Recombination Near Telomeres as Part of the Same Pathway", Genetics, vol. 183, pp. 441-451, 2009. http://dx.doi.org/10.1534/genetics.109.106674
- P. Baumann, and T.R. Cech, "Protection of Telomeres by the Ku Protein in Fission Yeast", Molecular Biology of the Cell, vol. 11, pp. 3265-3275, 2000. http://dx.doi.org/10.1091/mbc.11.10.3265
- O. Rog, K.M. Miller, M.G. Ferreira, and J.P. Cooper, "Sumoylation of RecQ Helicase Controls the Fate of Dysfunctional Telomeres", Molecular Cell, vol. 33, pp. 559-569, 2009. http://dx.doi.org/10.1016/j.molcel.2009.01.027
- N. Arat, and J.D. Griffith, "Human Rap1 Interacts Directly with Telomeric DNA and Regulates TRF2 Localization at the Telomere", Journal of Biological Chemistry, vol. 287, pp. 41583-41594, 2012. http://dx.doi.org/10.1074/jbc.M112.415984
- G.B. Celli, E.L. Denchi, and T. de Lange, "Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination", Nature Cell Biology, vol. 8, pp. 885-890, 2006. http://dx.doi.org/10.1038/ncb1444
- W. Palm, D. Hockemeyer, T. Kibe, and T. de Lange, "Functional Dissection of Human and Mouse POT1 Proteins", Molecular and Cellular Biology, vol. 29, pp. 471-482, 2009. http://dx.doi.org/10.1128/MCB.01352-08
- A. Sfeir, S. Kabir, M. van Overbeek, G.B. Celli, and T. de Lange, "Loss of Rap1 Induces Telomere Recombination in the Absence of NHEJ or a DNA Damage Signal", Science, vol. 327, pp. 1657-1661, 2010. http://dx.doi.org/10.1126/science.1185100
- J.D. Griffith, L. Comeau, S. Rosenfield, R.M. Stansel, A. Bianchi, H. Moss, and T. de Lange, "Mammalian Telomeres End in a Large Duplex Loop", Cell, vol. 97, pp. 503-514, 1999. http://dx.doi.org/10.1016/s0092-8674(00)80760-6
- Y. Doksani, J. Wu, T. de Lange, and X. Zhuang, "Super-Resolution Fluorescence Imaging of Telomeres Reveals TRF2-Dependent T-loop Formation", Cell, vol. 155, pp. 345-356, 2013. http://dx.doi.org/10.1016/j.cell.2013.09.048
- R.E. Verdun, and J. Karlseder, "The DNA Damage Machinery and Homologous Recombination Pathway Act Consecutively to Protect Human Telomeres", Cell, vol. 127, pp. 709-720, 2006. http://dx.doi.org/10.1016/j.cell.2006.09.034
- B.D. Bower, and J.D. Griffith, "TRF1 and TRF2 Differentially Modulate Rad51-Mediated Telomeric and Nontelomeric Displacement Loop Formation in Vitro", Biochemistry, vol. 53, pp. 5485-5495, 2014. http://dx.doi.org/10.1021/bi5006249
- G. Sarek, J. Vannier, S. Panier, J. Petrini, and S. Boulton, "TRF2 Recruits RTEL1 to Telomeres in S Phase to Promote T-Loop Unwinding", Molecular Cell, vol. 57, pp. 622-635, 2015. http://dx.doi.org/10.1016/j.molcel.2014.12.024
- T. de Lange, "How Telomeres Solve the End-Protection Problem", Science, vol. 326, pp. 948-952, 2009. http://dx.doi.org/10.1126/science.1170633
- J. Karlseder, K. Hoke, O.K. Mirzoeva, C. Bakkenist, M.B. Kastan, J.H.J. Petrini, and T.D. Lange, "The Telomeric Protein TRF2 Binds the ATM Kinase and Can Inhibit the ATM-Dependent DNA Damage Response", PLoS Biology, vol. 2, pp. e240, 2004. http://dx.doi.org/10.1371/journal.pbio.0020240
- R.C. Wang, A. Smogorzewska, and T. de Lange, "Homologous Recombination Generates T-Loop-Sized Deletions at Human Telomeres", Cell, vol. 119, pp. 355-368, 2004. http://dx.doi.org/10.1016/j.cell.2004.10.011
- Y. Liu, J. Masson, R. Shah, P. O'Regan, and S.C. West, "RAD51C Is Required for Holliday Junction Processing in Mammalian Cells", Science, vol. 303, pp. 243-246, 2004. http://dx.doi.org/10.1126/science.1093037
- A. Saint-Léger, M. Koelblen, L. Civitelli, A. Bah, N. Djerbi, M. Giraud-Panis, A. Londoño-Vallejo, F. Ascenzioni, and E. Gilson, "The basic N-terminal domain of TRF2 limits recombination endonuclease action at human telomeres", Cell Cycle, vol. 13, pp. 2469-2474, 2014. http://dx.doi.org/10.4161/cc.29422
- A.J. Cesare, C. Groff-Vindman, S.A. Compton, M.J. McEachern, and J.D. Griffith, "Telomere Loops and Homologous Recombination-Dependent Telomeric Circles in a Kluyveromyces lactis Telomere Mutant Strain", Molecular and Cellular Biology, vol. 28, pp. 20-29, 2008. http://dx.doi.org/10.1128/MCB.01122-07
- C. Lin, H. Chang, K. Wu, S. Tseng, C. Lin, C. Lin, and S. Teng, "Extrachromosomal Telomeric Circles Contribute to Rad52-, Rad50-, and Polymerase δ-Mediated Telomere-Telomere Recombination in Saccharomyces cerevisiae", Eukaryotic Cell, vol. 4, pp. 327-336, 2005. http://dx.doi.org/10.1128/EC.4.2.327-336.2005
- M. Larrivée, and R.J. Wellinger, "Telomerase- and capping-independent yeast survivors with alternate telomere states", Nature Cell Biology, vol. 8, pp. 741-747, 2006. http://dx.doi.org/10.1038/ncb1429
- A.J. Lustig, "Clues to catastrophic telomere loss in mammals from yeast telomere rapid deletion", Nature Reviews Genetics, vol. 4, pp. 916-923, 2003. http://dx.doi.org/10.1038/nrg1207
- H.A. Pickett, A.J. Cesare, R.L. Johnston, A.A. Neumann, and R.R. Reddel, "Control of telomere length by a trimming mechanism that involves generation of t-circles", The EMBO Journal, vol. 28, pp. 799-809, 2009. http://dx.doi.org/10.1038/emboj.2009.42
- B. Li, and A.J. Lustig, "A novel mechanism for telomere size control in Saccharomyces cerevisiae.", Genes & Development, vol. 10, pp. 1310-1326, 1996. http://dx.doi.org/10.1101/gad.10.11.1310
- M. Bucholc, Y. Park, and A.J. Lustig, "Intrachromatid Excision of Telomeric DNA as a Mechanism for Telomere Size Control in Saccharomyces cerevisiae", Molecular and Cellular Biology, vol. 21, pp. 6559-6573, 2001. http://dx.doi.org/10.1128/mcb.21.19.6559-6573.2001
- L.H. Bechard, N. Jamieson, and M.J. McEachern, "Recombination Can Cause Telomere Elongations as Well as Truncations Deep within Telomeres in Wild-Type Kluyveromyces lactis Cells", Eukaryotic Cell, vol. 10, pp. 226-236, 2011. http://dx.doi.org/10.1128/EC.00209-10
- J.M. Watson, and D.E. Shippen, "Telomere Rapid Deletion Regulates Telomere Length in Arabidopsis thaliana", Molecular and Cellular Biology, vol. 27, pp. 1706-1715, 2007. http://dx.doi.org/10.1128/MCB.02059-06
- H.A. Pickett, J.D. Henson, A.Y.M. Au, A.A. Neumann, and R.R. Reddel, "Normal mammalian cells negatively regulate telomere length by telomere trimming", Human Molecular Genetics, vol. 20, pp. 4684-4692, 2011. http://dx.doi.org/10.1093/hmg/ddr402
- J.T.P. Yeeles, J. Poli, K.J. Marians, and P. Pasero, "Rescuing Stalled or Damaged Replication Forks", Cold Spring Harbor Perspectives in Biology, vol. 5, pp. a012815-a012815, 2013. http://dx.doi.org/10.1101/cshperspect.a012815
- A.S. Ivessa, J. Zhou, V.P. Schulz, E.K. Monson, and V.A. Zakian, "Saccharomyces Rrm3p, a 5′ to 3′ DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA", Genes & Development, vol. 16, pp. 1383-1396, 2002. http://dx.doi.org/10.1101/gad.982902
- K.M. Miller, O. Rog, and J.P. Cooper, "Semi-conservative DNA replication through telomeres requires Taz1", Nature, vol. 440, pp. 824-828, 2006. http://dx.doi.org/10.1038/nature04638
- A. Sfeir, S.T. Kosiyatrakul, D. Hockemeyer, S.L. MacRae, J. Karlseder, C.L. Schildkraut, and T. de Lange, "Mammalian Telomeres Resemble Fragile Sites and Require TRF1 for Efficient Replication", Cell, vol. 138, pp. 90-103, 2009. http://dx.doi.org/10.1016/j.cell.2009.06.021
- S. Badie, J.M. Escandell, P. Bouwman, A.R. Carlos, M. Thanasoula, M.M. Gallardo, A. Suram, I. Jaco, J. Benitez, U. Herbig, M.A. Blasco, J. Jonkers, and M. Tarsounas, "BRCA2 acts as a RAD51 loader to facilitate telomere replication and capping", Nature Structural & Molecular Biology, vol. 17, pp. 1461-1469, 2010. http://dx.doi.org/10.1038/nsmb.1943
- M. Tarsounas, P. Muñoz, A. Claas, P.G. Smiraldo, D.L. Pittman, M.A. Blasco, and S.C. West, "Telomere Maintenance Requires the RAD51D Recombination/Repair Protein", Cell, vol. 117, pp. 337-347, 2004. http://dx.doi.org/10.1016/s0092-8674(04)00337-x
- I. Jaco, P. Muñoz, F. Goytisolo, J. Wesoly, S. Bailey, G. Taccioli, and M.A. Blasco, "Role of Mammalian Rad54 in Telomere Length Maintenance", Molecular and Cellular Biology, vol. 23, pp. 5572-5580, 2003. http://dx.doi.org/10.1128/mcb.23.16.5572-5580.2003
- H. Gaillard, E. Herrera-Moyano, and A. Aguilera, "Transcription-Associated Genome Instability", Chemical Reviews, vol. 113, pp. 8638-8661, 2013. http://dx.doi.org/10.1021/cr400017y
- B. Balk, M. Dees, K. Bender, and B. Luke, "The differential processing of telomeres in response to increased telomeric transcription and RNA–DNA hybrid accumulation", RNA Biology, vol. 11, pp. 95-100, 2014. http://dx.doi.org/10.4161/rna.27798
- B. Balk, A. Maicher, M. Dees, J. Klermund, S. Luke-Glaser, K. Bender, and B. Luke, "Telomeric RNA-DNA hybrids affect telomere-length dynamics and senescence", Nature Structural & Molecular Biology, vol. 20, pp. 1199-1205, 2013. http://dx.doi.org/10.1038/nsmb.2662
- V. Pfeiffer, J. Crittin, L. Grolimund, and J. Lingner, "The THO complex component Thp2 counteracts telomeric R-loops and telomere shortening", The EMBO Journal, vol. 32, pp. 2861-2871, 2013. http://dx.doi.org/10.1038/emboj.2013.217
- T. Yu, Y. Kao, and J. Lin, "Telomeric transcripts stimulate telomere recombination to suppress senescence in cells lacking telomerase", Proceedings of the National Academy of Sciences, vol. 111, pp. 3377-3382, 2014. http://dx.doi.org/10.1073/pnas.1307415111
- S.C. Teng, and V.A. Zakian, "Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae.", Molecular and cellular biology, 1999. http://www.ncbi.nlm.nih.gov/pubmed/10567534
- J.R. Lydeard, S. Jain, M. Yamaguchi, and J.E. Haber, "Break-induced replication and telomerase-independent telomere maintenance require Pol32", Nature, vol. 448, pp. 820-823, 2007. http://dx.doi.org/10.1038/nature06047
- J.M. Dewar, and D. Lydall, "Pif1- and Exo1-dependent nucleases coordinate checkpoint activation following telomere uncapping", The EMBO Journal, vol. 29, pp. 4020-4034, 2010. http://dx.doi.org/10.1038/emboj.2010.267
- Y. Hu, H. Tang, N. Liu, X. Tong, W. Dang, Y. Duan, X. Fu, Y. Zhang, J. Peng, F. Meng, and J. Zhou, "Telomerase-Null Survivor Screening Identifies Novel Telomere Recombination Regulators", PLoS Genetics, vol. 9, pp. e1003208, 2013. http://dx.doi.org/10.1371/journal.pgen.1003208
- M.A. Wilson, Y. Kwon, Y. Xu, W. Chung, P. Chi, H. Niu, R. Mayle, X. Chen, A. Malkova, P. Sung, and G. Ira, "Pif1 helicase and Polδ promote recombination-coupled DNA synthesis via bubble migration", Nature, vol. 502, pp. 393-396, 2013. http://dx.doi.org/10.1038/nature12585
- A.P. Davis, and L.S. Symington, "RAD51-Dependent Break-Induced Replication in Yeast", Molecular and Cellular Biology, vol. 24, pp. 2344-2351, 2004. http://dx.doi.org/10.1128/mcb.24.6.2344-2351.2004
- L. Signon, A. Malkova, M.L. Naylor, H. Klein, and J.E. Haber, "Genetic Requirements for RAD51- andRAD54-Independent Break-Induced Replication Repair of a Chromosomal Double-Strand Break", Molecular and Cellular Biology, vol. 21, pp. 2048-2056, 2001. http://dx.doi.org/10.1128/MCB.21.6.2048-2056.2001
- Q. Chen, A. Ijpma, and C.W. Greider, "Two Survivor Pathways That Allow Growth in the Absence of Telomerase Are Generated by Distinct Telomere Recombination Events", Molecular and Cellular Biology, vol. 21, pp. 1819-1827, 2001. http://dx.doi.org/10.1128/MCB.21.5.1819-1827.2001
- P. Huang, F.E. Pryde, D. Lester, R.L. Maddison, R.H. Borts, I.D. Hickson, and E.J. Louis, "SGS1 is required for telomere elongation in the absence of telomerase", Current Biology, vol. 11, pp. 125-129, 2001. http://dx.doi.org/10.1016/s0960-9822(01)00021-5
- F. Johnson, "The Saccharomyces cerevisiae WRN homolog Sgs1p participates in telomere maintenance in cells lacking telomerase", The EMBO Journal, vol. 20, pp. 905-913, 2001. http://dx.doi.org/10.1093/emboj/20.4.905
- L. Maringele, "EXO1 Plays a Role in Generating Type I and Type II Survivors in Budding Yeast", Genetics, vol. 166, pp. 1641-1649, 2004. http://dx.doi.org/10.1534/genetics.166.4.1641
- A.A. Bertuch, "EXO1 Contributes to Telomere Maintenance in Both Telomerase-Proficient and Telomerase-Deficient Saccharomyces cerevisiae", Genetics, vol. 166, pp. 1651-1659, 2004. http://dx.doi.org/10.1534/genetics.166.4.1651
- K.A. Bernstein, E. Shor, I. Sunjevaric, M. Fumasoni, R.C. Burgess, M. Foiani, D. Branzei, and R. Rothstein, "Sgs1 function in the repair of DNA replication intermediates is separable from its role in homologous recombinational repair", The EMBO Journal, vol. 28, pp. 915-925, 2009. http://dx.doi.org/10.1038/emboj.2009.28
- K.A. Bernstein, E.P. Mimitou, M.J. Mihalevic, H. Chen, I. Sunjaveric, L.S. Symington, and R. Rothstein, "Resection Activity of the Sgs1 Helicase Alters the Affinity of DNA Ends for Homologous Recombination Proteins inSaccharomyces cerevisiae", Genetics, vol. 195, pp. 1241-1251, 2013. http://dx.doi.org/10.1534/genetics.113.157370
- J. Hardy, D. Churikov, V. Géli, and M. Simon, "Sgs1 and Sae2 promote telomere replication by limiting accumulation of ssDNA", Nature Communications, vol. 5, 2014. http://dx.doi.org/10.1038/ncomms6004
- J.R. Lydeard, Z. Lipkin-Moore, S. Jain, V.V. Eapen, and J.E. Haber, "Sgs1 and Exo1 Redundantly Inhibit Break-Induced Replication and De Novo Telomere Addition at Broken Chromosome Ends", PLoS Genetics, vol. 6, pp. e1000973, 2010. http://dx.doi.org/10.1371/journal.pgen.1000973
- V.A. Marrero, and L.S. Symington, "Extensive DNA End Processing by Exo1 and Sgs1 Inhibits Break-Induced Replication", PLoS Genetics, vol. 6, pp. e1001007, 2010. http://dx.doi.org/10.1371/journal.pgen.1001007
- T. Costelloe, R. Louge, N. Tomimatsu, B. Mukherjee, E. Martini, B. Khadaroo, K. Dubois, W.W. Wiegant, A. Thierry, S. Burma, H. van Attikum, and B. Llorente, "The yeast Fun30 and human SMARCAD1 chromatin remodellers promote DNA end resection", Nature, vol. 489, pp. 581-584, 2012. http://dx.doi.org/10.1038/nature11353
- Y. Tsai, S. Tseng, S. Chang, C. Lin, and S. Teng, "Involvement of Replicative Polymerases, Tel1p, Mec1p, Cdc13p, and the Ku Complex in Telomere-Telomere Recombination", Molecular and Cellular Biology, vol. 22, pp. 5679-5687, 2002. http://dx.doi.org/10.1128/mcb.22.16.5679-5687.2002
- N. Grandin, and M. Charbonneau, "Mitotic Cyclins Regulate Telomeric Recombination in Telomerase-Deficient Yeast Cells", Molecular and Cellular Biology, vol. 23, pp. 9162-9177, 2003. http://dx.doi.org/10.1128/mcb.23.24.9162-9177.2003
- Y. Chen, C. Yang, R. Li, R. Zeng, and J. Zhou, "Def1p Is Involved in Telomere Maintenance in Budding Yeast", Journal of Biological Chemistry, vol. 280, pp. 24784-24791, 2005. http://dx.doi.org/10.1074/jbc.M413562200
- B.L. Pike, and J. Heierhorst, "Mdt1 Facilitates Efficient Repair of Blocked DNA Double-Strand Breaks and Recombinational Maintenance of Telomeres", Molecular and Cellular Biology, vol. 27, pp. 6532-6545, 2007. http://dx.doi.org/10.1128/MCB.00471-07
- F. Meng, X. Chen, Y. Hu, H. Tang, W. Dang, and J. Zhou, "Sua5p is required for telomere recombination in Saccharomyces cerevisiae", Cell Research, vol. 20, pp. 495-498, 2010. http://dx.doi.org/10.1038/cr.2010.34
- S. Teng, J. Chang, B. McCowan, and V.A. Zakian, "Telomerase-Independent Lengthening of Yeast Telomeres Occurs by an Abrupt Rad50p-Dependent, Rif-Inhibited Recombinational Process", Molecular Cell, vol. 6, pp. 947-952, 2000. http://dx.doi.org/10.1016/s1097-2765(05)00094-8
- K.R. Straatman, and E.J. Louis, "Localization of telomeres and telomere-associated proteins in telomerase-negative Saccharomyces cerevisiae", Chromosome Research, vol. 15, 2007. http://dx.doi.org/10.1007/s10577-007-1178-2
- E.J. Louis, E.S. Naumova, A. Lee, G. Naumov, and J.E. Haber, "The chromosome end in yeast: its mosaic nature and influence on recombinational dynamics.", Genetics, 1994. http://www.ncbi.nlm.nih.gov/pubmed/8005434
- D. Churikov, F. Charifi, M. Simon, and V. Géli, "Rad59-Facilitated Acquisition of Y′ Elements by Short Telomeres Delays the Onset of Senescence", PLoS Genetics, vol. 10, pp. e1004736, 2014. http://dx.doi.org/10.1371/journal.pgen.1004736
- M. Yamada, N. Hayatsu, A. Matsuura, and F. Ishikawa, "Y′-Help1, a DNA Helicase Encoded by the Yeast Subtelomeric Y′ Element, Is Induced in Survivors Defective for Telomerase", Journal of Biological Chemistry, vol. 273, pp. 33360-33366, 1998. http://dx.doi.org/10.1074/jbc.273.50.33360
- M.J. McEachern, and J.E. Haber, "Break-Induced Replication and Recombinational Telomere Elongation in Yeast", Annual Review of Biochemistry, vol. 75, pp. 111-135, 2006. http://dx.doi.org/10.1146/annurev.biochem.74.082803.133234
- M.J. McEachern, and E.H. Blackburn, "Cap-prevented recombination between terminal telomeric repeat arrays (telomere CPR) maintains telomeres in Kluyveromyces lactis lacking telomerase.", Genes & Development, vol. 10, pp. 1822-1834, 1996. http://dx.doi.org/10.1101/gad.10.14.1822
- S. Natarajan, and M.J. McEachern, "Recombinational Telomere Elongation Promoted by DNA Circles", Molecular and Cellular Biology, vol. 22, pp. 4512-4521, 2002. http://dx.doi.org/10.1128/mcb.22.13.4512-4521.2002
- C. Groff-Vindman, A.J. Cesare, S. Natarajan, J.D. Griffith, and M.J. McEachern, "Recombination at Long Mutant Telomeres Produces Tiny Single- and Double-Stranded Telomeric Circles", Molecular and Cellular Biology, vol. 25, pp. 4406-4412, 2005. http://dx.doi.org/10.1128/MCB.25.11.4406-4412.2005
- A.J. Cesare, and J.D. Griffith, "Telomeric DNA in ALT Cells Is Characterized by Free Telomeric Circles and Heterogeneous t-Loops", Molecular and Cellular Biology, vol. 24, pp. 9948-9957, 2004. http://dx.doi.org/10.1128/MCB.24.22.9948-9957.2004
- P. Dehé, and J.P. Cooper, "Fission yeast telomeres forecast the end of the crisis", FEBS Letters, vol. 584, pp. 3725-3733, 2010. http://dx.doi.org/10.1016/j.febslet.2010.07.045
- T.M. Nakamura, J.P. Cooper, and T.R. Cech, "Two Modes of Survival of Fission Yeast Without Telomerase", Science, vol. 282, pp. 493-496, 1998. http://dx.doi.org/10.1126/science.282.5388.493
- L. Subramanian, B.A. Moser, and T.M. Nakamura, "Recombination-Based Telomere Maintenance Is Dependent on Tel1-MRN and Rap1 and Inhibited by Telomerase, Taz1, and Ku in Fission Yeast", Molecular and Cellular Biology, vol. 28, pp. 1443-1455, 2008. http://dx.doi.org/10.1128/MCB.01614-07
- N. Grandin, and M. Charbonneau, "Telomerase- and Rad52-Independent Immortalization of Budding Yeast by an Inherited-Long-Telomere Pathway of Telomeric Repeat Amplification", Molecular and Cellular Biology, vol. 29, pp. 965-985, 2009. http://dx.doi.org/10.1128/MCB.00817-08
- C. LeBel, E. Rosonina, D.C.F. Sealey, F. Pryde, D. Lydall, L. Maringele, and L.A. Harrington, "Telomere Maintenance and Survival in Saccharomyces cerevisiae in the Absence of Telomerase and RAD52", Genetics, vol. 182, pp. 671-684, 2009. http://dx.doi.org/10.1534/genetics.109.102939
- B.A. Ozenberger, and G.S. Roeder, "A unique pathway of double-strand break repair operates in tandemly repeated genes.", Molecular and cellular biology, 1991. http://www.ncbi.nlm.nih.gov/pubmed/1996088
- L. Maringele, and D. Lydall, "Telomerase- and recombination-independent immortalization of budding yeast", Genes & Development, vol. 18, pp. 2663-2675, 2004. http://dx.doi.org/10.1101/gad.316504
- J.Y. Lee, J.L. Mogen, A. Chavez, and F.B. Johnson, "Sgs1 RecQ Helicase Inhibits Survival of Saccharomyces cerevisiae Cells Lacking Telomerase and Homologous Recombination", Journal of Biological Chemistry, vol. 283, pp. 29847-29858, 2008. http://dx.doi.org/10.1074/jbc.M804760200
- C.L. Fasching, K. Bower, and R.R. Reddel, "Telomerase-Independent Telomere Length Maintenance in the Absence of Alternative Lengthening of Telomeres–Associated Promyelocytic Leukemia Bodies", Cancer Research, vol. 65, pp. 2722-2729, 2005. http://dx.doi.org/10.1158/0008-5472.CAN-04-2881
- R.A. Marciniak, D. Cavazos, R. Montellano, Q. Chen, L. Guarente, and F.B. Johnson, "A Novel Telomere Structure in a Human Alternative Lengthening of Telomeres Cell Line", Cancer Research, vol. 65, pp. 2730-2737, 2005. http://dx.doi.org/10.1158/0008-5472.CAN-04-2888
- T.M. Bryan, A. Englezou, J. Gupta, S. Bacchetti, and R.R. Reddel, "Telomere elongation in immortal human cells without detectable telomerase activity.", The EMBO journal, 1995. http://www.ncbi.nlm.nih.gov/pubmed/7556065
- J.D. Henson, Y. Cao, L.I. Huschtscha, A.C. Chang, A.Y.M. Au, H.A. Pickett, and R.R. Reddel, "DNA C-circles are specific and quantifiable markers of alternative-lengthening-of-telomeres activity", Nature Biotechnology, vol. 27, pp. 1181-1185, 2009. http://dx.doi.org/10.1038/nbt.1587
- W. Jiang, Z. Zhong, J.D. Henson, A.A. Neumann, A.C. Chang, and R.R. Reddel, "Suppression of Alternative Lengthening of Telomeres by Sp100-Mediated Sequestration of the MRE11/RAD50/NBS1 Complex", Molecular and Cellular Biology, vol. 25, pp. 2708-2721, 2005. http://dx.doi.org/10.1128/MCB.25.7.2708-2721.2005
- Z. Zhong, W. Jiang, A.J. Cesare, A.A. Neumann, R. Wadhwa, and R.R. Reddel, "Disruption of Telomere Maintenance by Depletion of the MRE11/RAD50/NBS1 Complex in Cells That Use Alternative Lengthening of Telomeres", Journal of Biological Chemistry, vol. 282, pp. 29314-29322, 2007. http://dx.doi.org/10.1074/jbc.M701413200
- S. Bhattacharyya, J. Keirsey, B. Russell, J. Kavecansky, K. Lillard-Wetherell, K. Tahmaseb, J.J. Turchi, and J. Groden, "Telomerase-associated Protein 1, HSP90, and Topoisomerase IIα Associate Directly with the BLM Helicase in Immortalized Cells Using ALT and Modulate Its Helicase Activity Using Telomeric DNA Substrates", Journal of Biological Chemistry, vol. 284, pp. 14966-14977, 2009. http://dx.doi.org/10.1074/jbc.M900195200
- A.R.S. Gocha, S. Acharya, and J. Groden, "WRN Loss Induces Switching of Telomerase-Independent Mechanisms of Telomere Elongation", PLoS ONE, vol. 9, pp. e93991, 2014. http://dx.doi.org/10.1371/journal.pone.0093991
- T.R. Yeager, A.A. Neumann, A. Englezou, L.I. Huschtscha, J.R. Noble, and R.R. Reddel, "Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body.", Cancer research, 1999. http://www.ncbi.nlm.nih.gov/pubmed/10485449
- A. Nabetani, and F. Ishikawa, "Alternative lengthening of telomeres pathway: Recombination-mediated telomere maintenance mechanism in human cells", Journal of Biochemistry, vol. 149, pp. 5-14, 2010. http://dx.doi.org/10.1093/jb/mvq119
- I. Chung, S. Osterwald, K.I. Deeg, and K. Rippe, "PML body meets telomere", Nucleus, vol. 3, pp. 263-275, 2012. http://dx.doi.org/10.4161/nucl.20326
- O.E. Bechter, Y. Zou, W. Walker, W.E. Wright, and J.W. Shay, "Telomeric Recombination in Mismatch Repair Deficient Human Colon Cancer Cells after Telomerase Inhibition", Cancer Research, vol. 64, pp. 3444-3451, 2004. http://dx.doi.org/10.1158/0008-5472.CAN-04-0323
- J.A. Londoño-Vallejo, H. Der-Sarkissian, L. Cazes, S. Bacchetti, and R.R. Reddel, "Alternative Lengthening of Telomeres Is Characterized by High Rates of Telomeric Exchange", Cancer Research, vol. 64, pp. 2324-2327, 2004. http://dx.doi.org/10.1158/0008-5472.can-03-4035
- C.A. Lovejoy, W. Li, S. Reisenweber, S. Thongthip, J. Bruno, T. de Lange, S. De, J.H.J. Petrini, P.A. Sung, M. Jasin, J. Rosenbluh, Y. Zwang, B.A. Weir, C. Hatton, E. Ivanova, L. Macconaill, M. Hanna, W.C. Hahn, N.F. Lue, R.R. Reddel, Y. Jiao, K. Kinzler, B. Vogelstein, N. Papadopoulos, A.K. Meeker, and . , "Loss of ATRX, Genome Instability, and an Altered DNA Damage Response Are Hallmarks of the Alternative Lengthening of Telomeres Pathway", PLoS Genetics, vol. 8, pp. e1002772, 2012. http://dx.doi.org/10.1371/journal.pgen.1002772
- R.L. Flynn, K.E. Cox, M. Jeitany, H. Wakimoto, A.R. Bryll, N.J. Ganem, F. Bersani, J.R. Pineda, M.L. Suvà, C.H. Benes, D.A. Haber, F.D. Boussin, and L. Zou, "Alternative lengthening of telomeres renders cancer cells hypersensitive to ATR inhibitors", Science, vol. 347, pp. 273-277, 2015. http://dx.doi.org/10.1126/science.1257216
- R.L. Flynn, R.C. Centore, R.J. O’Sullivan, R. Rai, A. Tse, Z. Songyang, S. Chang, J. Karlseder, and L. Zou, "TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA", Nature, vol. 471, pp. 532-536, 2011. http://dx.doi.org/10.1038/nature09772
- L.J. Ng, J.E. Cropley, H.A. Pickett, R.R. Reddel, and C.M. Suter, "Telomerase activity is associated with an increase in DNA methylation at the proximal subtelomere and a reduction in telomeric transcription", Nucleic Acids Research, vol. 37, pp. 1152-1159, 2009. http://dx.doi.org/10.1093/nar/gkn1030
- R.J. O'Sullivan, N. Arnoult, D.H. Lackner, L. Oganesian, C. Haggblom, A. Corpet, G. Almouzni, and J. Karlseder, "Rapid induction of alternative lengthening of telomeres by depletion of the histone chaperone ASF1", Nature Structural & Molecular Biology, vol. 21, pp. 167-174, 2014. http://dx.doi.org/10.1038/nsmb.2754
- P. Drané, K. Ouararhni, A. Depaux, M. Shuaib, and A. Hamiche, "The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3", Genes & Development, vol. 24, pp. 1253-1265, 2010. http://dx.doi.org/10.1101/gad.566910
- A.D. Goldberg, L.A. Banaszynski, K. Noh, P.W. Lewis, S.J. Elsaesser, S. Stadler, S. Dewell, M. Law, X. Guo, X. Li, D. Wen, A. Chapgier, R.C. DeKelver, J.C. Miller, Y. Lee, E.A. Boydston, M.C. Holmes, P.D. Gregory, J.M. Greally, S. Rafii, C. Yang, P.J. Scambler, D. Garrick, R.J. Gibbons, D.R. Higgs, I.M. Cristea, F.D. Urnov, D. Zheng, and C.D. Allis, "Distinct Factors Control Histone Variant H3.3 Localization at Specific Genomic Regions", Cell, vol. 140, pp. 678-691, 2010. http://dx.doi.org/10.1016/j.cell.2010.01.003
- M.J. Law, K.M. Lower, H.P. Voon, J.R. Hughes, D. Garrick, V. Viprakasit, M. Mitson, M. De Gobbi, M. Marra, A. Morris, A. Abbott, S.P. Wilder, S. Taylor, G.M. Santos, J. Cross, H. Ayyub, S. Jones, J. Ragoussis, D. Rhodes, I. Dunham, D.R. Higgs, and R.J. Gibbons, "ATR-X Syndrome Protein Targets Tandem Repeats and Influences Allele-Specific Expression in a Size-Dependent Manner", Cell, vol. 143, pp. 367-378, 2010. http://dx.doi.org/10.1016/j.cell.2010.09.023
- P.W. Lewis, S.J. Elsaesser, K. Noh, S.C. Stadler, and C.D. Allis, "Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres", Proceedings of the National Academy of Sciences, vol. 107, pp. 14075-14080, 2010. http://dx.doi.org/10.1073/pnas.1008850107
- L.H. Wong, J.D. McGhie, M. Sim, M.A. Anderson, S. Ahn, R.D. Hannan, A.J. George, K.A. Morgan, J.R. Mann, and K.A. Choo, "ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells", Genome Research, vol. 20, pp. 351-360, 2010. http://dx.doi.org/10.1101/gr.101477.109
- C.M. Heaphy, R.F. de Wilde, Y. Jiao, A.P. Klein, B.H. Edil, C. Shi, C. Bettegowda, F.J. Rodriguez, C.G. Eberhart, S. Hebbar, G.J. Offerhaus, R. McLendon, B.A. Rasheed, Y. He, H. Yan, D.D. Bigner, S.M. Oba-Shinjo, S.K.N. Marie, G.J. Riggins, K.W. Kinzler, B. Vogelstein, R.H. Hruban, A. Maitra, N. Papadopoulos, and A.K. Meeker, "Altered Telomeres in Tumors with ATRX and DAXX Mutations", Science, vol. 333, pp. 425-425, 2011. http://dx.doi.org/10.1126/science.1207313
- J. Schwartzentruber, A. Korshunov, X. Liu, D.T.W. Jones, E. Pfaff, K. Jacob, D. Sturm, A.M. Fontebasso, D.K. Quang, M. Tönjes, V. Hovestadt, S. Albrecht, M. Kool, A. Nantel, C. Konermann, A. Lindroth, N. Jäger, T. Rausch, M. Ryzhova, J.O. Korbel, T. Hielscher, P. Hauser, M. Garami, A. Klekner, L. Bognar, M. Ebinger, M.U. Schuhmann, W. Scheurlen, A. Pekrun, M.C. Frühwald, W. Roggendorf, C. Kramm, M. Dürken, J. Atkinson, P. Lepage, A. Montpetit, M. Zakrzewska, K. Zakrzewski, P.P. Liberski, Z. Dong, P. Siegel, A.E. Kulozik, M. Zapatka, A. Guha, D. Malkin, J. Felsberg, G. Reifenberger, A. von Deimling, K. Ichimura, V.P. Collins, H. Witt, T. Milde, O. Witt, C. Zhang, P. Castelo-Branco, P. Lichter, D. Faury, U. Tabori, C. Plass, J. Majewski, S.M. Pfister, and N. Jabado, "Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma", Nature, vol. 482, pp. 226-231, 2012. http://dx.doi.org/10.1038/nature10833
- K. Bower, C.E. Napier, S.L. Cole, R.A. Dagg, L.M.S. Lau, E.L. Duncan, E.L. Moy, and R.R. Reddel, "Loss of Wild-Type ATRX Expression in Somatic Cell Hybrids Segregates with Activation of Alternative Lengthening of Telomeres", PLoS ONE, vol. 7, pp. e50062, 2012. http://dx.doi.org/10.1371/journal.pone.0050062
- S. Le, C. Davis, J.B. Konopka, and R. Sternglanz, "Two new S-phase-specific genes from Saccharomyces cerevisiae.", Yeast (Chichester, England), 1997. http://www.ncbi.nlm.nih.gov/pubmed/9290207
- J.K. Tyler, C.R. Adams, S. Chen, R. Kobayashi, R.T. Kamakaka, and J.T. Kadonaga, "The RCAF complex mediates chromatin assembly during DNA replication and repair", Nature, vol. 402, pp. 555-560, 1999. http://dx.doi.org/10.1038/990147
- H. Tagami, D. Ray-Gallet, G. Almouzni, and Y. Nakatani, "Histone H3.1 and H3.3 Complexes Mediate Nucleosome Assembly Pathways Dependent or Independent of DNA Synthesis", Cell, vol. 116, pp. 51-61, 2004. http://dx.doi.org/10.1016/s0092-8674(03)01064-x
- X. Fu, Y. Duan, Y. Liu, C. Cai, F. Meng, and J. Zhou, "Telomere Recombination Preferentially Occurs at Short Telomeres in Telomerase-Null Type II Survivors", PLoS ONE, vol. 9, pp. e90644, 2014. http://dx.doi.org/10.1371/journal.pone.0090644
- M.J. McEachern, and S. Iyer, "Short Telomeres in Yeast Are Highly Recombinogenic", Molecular Cell, vol. 7, pp. 695-704, 2001. http://dx.doi.org/10.1016/s1097-2765(01)00215-5
- Z. Topcu, K. Nickles, C. Davis, and M.J. McEachern, "Abrupt disruption of capping and a single source for recombinationally elongated telomeres in Kluyveromyces lactis", Proceedings of the National Academy of Sciences, vol. 102, pp. 3348-3353, 2005. http://dx.doi.org/10.1073/pnas.0408770102
- B.S. Singer, L. Gold, P. Gauss, and D.H. Doherty, "Determination of the amount of homology required for recombination in bacteriophage T4", Cell, vol. 31, pp. 25-33, 1982. http://dx.doi.org/10.1016/0092-8674(82)90401-9
- V.M. Watt, C.J. Ingles, M.S. Urdea, and W.J. Rutter, "Homology requirements for recombination in Escherichia coli.", Proceedings of the National Academy of Sciences, vol. 82, pp. 4768-4772, 1985. http://dx.doi.org/10.1073/pnas.82.14.4768
- P. Shen, and H.V. Huang, "Homologous recombination in Escherichia coli: dependence on substrate length and homology.", Genetics, 1986. http://www.ncbi.nlm.nih.gov/pubmed/3007275
- S. Jinks-Robertson, M. Michelitch, and S. Ramcharan, "Substrate length requirements for efficient mitotic recombination in Saccharomyces cerevisiae.", Molecular and cellular biology, 1993. http://www.ncbi.nlm.nih.gov/pubmed/8321201
- J. Rubnitz, and S. Subramani, "The minimum amount of homology required for homologous recombination in mammalian cells.", Molecular and cellular biology, 1984. http://www.ncbi.nlm.nih.gov/pubmed/6096689
- R.M. Liskay, A. Letsou, and J.L. Stachelek, "Homology requirement for efficient gene conversion between duplicated chromosomal sequences in mammalian cells.", Genetics, 1987. http://www.ncbi.nlm.nih.gov/pubmed/3557108
ACKNOWLEDGMENTS
We thank Evert-Jan Uringa and Brian Luke for constructive comments on the manuscript. Work in the Chang lab is supported by a Vidi grant (to MC) from the Netherlands Organization for Scientific Research (NWO).
COPYRIGHT
© 2015

The many facets of homologous recombination at telomeres by Clémence Claussin and Michael Chang is licensed under a Creative Commons Attribution 4.0 International License.