Research Articles:
Microbial Cell, Vol. 7, No. 6, pp. 146 - 159; doi: 10.15698/mic2020.06.719
Broad-spectrum antifungal activities and mechanism of drimane sesquiterpenoids
1 Department of Chemistry, 1212 Mid Campus Drive North, Kansas State University, Manhattan, KS 66506 USA.
2 Division of Biology, 1717 Claflin Road, Kansas State University, Manhattan, KS 66506 USA.
3 Faculty of Pharmaceutical Sciences, University of British Columbia,Vancouver, BC Canada V6T 1Z3.
Keywords: drimenol, synthesis, Candida albicans, Candida auris, Saccharomyces cerevisiae, fungicidal antifungal, novel mechanism of action, Crk1 kinase associated.
Abbreviations:
CLSI – clinical laboratory standard institute,
DMSO – dimethyl sulfoxide,
ER – endoplasmic reticulum,
FLU – fluconazole,
MOA – mechanism of action,
ROS – reactive oxygen species.
Received originally: 10/11/2020 Received in revised form: 29/02/2020
Accepted: 04/03/2020
Published: 12/03/2020
Correspondence:
Govindsamy Vediyappan, 1717 Claflin Road, Division of Biology, Kansas State University, Manhattan KS 66506 Tel. 785 532 6662, Fax. 785 532 6653; gvediyap@ksu.edu
Conflict of interest statement: A US patent (US 8,980,951 B2) on synthetic drimenol was approved in 2015 to Kansas State University Re-search Foundation (KSURF) with authors GV and DHH.
Please cite this article as: Edruce Edouarzin, Connor Horn, Anuja Paudyal, Cunli Zhang, Jianyu Lu, Zongbo Tong, Guri Giaever, Corey Nislow, Raja Veera-pandian, Duy H. Hua and Govindsamy Vediyappan (2020). Broad-spectrum antifungal activities and mechanism of drimane sesquiterpenoids. Microbial Cell 7(6): 146-159. doi: 10.15698/mic2020.06.719
Abstract
Eight drimane sesquiterpenoids including (-)-drimenol and (+)-albicanol were synthesized from (+)-sclareolide and evaluated for their antifungal activities. Three compounds, (-)-drimenol, (+)-albicanol, and (1R,2R,4aS,8aS)-2-hydroxy-2,5,5,8a-tetramethyl-decahydronaphthalene-1-carbaldehyde (4) showed strong activity against C. albicans. (-)-Drimenol, the strongest inhibitor of the three, (at concentrations of 8 – 64 µg/ml, causing 100% death of various fungi), acts not only against C. albicans in a fungicidal manner, but also inhibits other fungi such as Aspergillus, Cryptococcus, Pneumocystis, Blastomyces, Saksenaea and fluconazole resistant strains of C. albicans, C. glabrata, C. krusei, C. parapsilosis and C. auris. These observations suggest that drimenol is a broad-spectrum antifungal agent. At a high concentration (100 μg/ml) drimenol caused rupture of the fungal cell wall/membrane. In a nematode model of C. albicans infection, drimenol rescued the worms from C. albicans-mediated death, indicating drimenol is tolerable and bioactive in metazoans. Genome-wide fitness profiling assays of both S. cerevisiae (nonessential homozygous and essential heterozygous) and C. albicans (Tn-insertion mutants) collections revealed putative genes and pathways affected by drimenol. Using a C. albicans mutant spot assay, the Crk1 kinase associated gene products, Ret2, Cdc37, and orf19.759, orf19.1672, and orf19.4382 were revealed to be involved in drimenol’s mechanism of action. The three orfs identified in this study are novel and appear to be linked with Crk1 function. Further, computational modeling results suggest possible modifications of the structure of drimenol, including the A ring, for improving the antifungal activity.
INTRODUCTION
Fungi have emerged in the last two decades as major causes of human disease. Candida albicans is a major fungal pathogen affecting humans of all ages and is the fourth leading cause of nosocomial bloodstream infections in the US [1]. C. albicans is the most frequently found fungal pathogen in humans and costs the US health care system around $3 billion annually due to treatment costs and lost productivity [2][3]. According to a recent report the total global costs due to productivity loss caused by Candidiasis in women was estimated to be over $14 billion in 2010 [4]. C. albicans, a polymorphic fungus, exists as yeast, pseudohyphal and hyphal forms, with each contributing to its virulence. While the yeast form is essential for dissemination, the hyphal form is critical for invasion of cells, immune evasion, and biofilm formation. Furthermore, the ability to switch between forms is also essential for pathogenicity.
–
C. albicans and other Candida spp. cause mucosal and disseminate invasive candidiasis, especially among patients who are immunocompromised or hospitalized with serious underlying diseases. The overall mortality of invasive diseases caused by Candida spp. and Aspergillus spp. is around 50% [1][5]. While there are more than 150 species of Candida, about 15 species are recognized as frequent human pathogens [5][6]. Some of them are: C. albicans, C. glabrata, C. krusei, C. tropicalis and C. parapsilosis. Among these, C. albicans is by far the most common species isolated from humans and is a frequent denizen of the oropharynx, mucousal surfaces, gastrointestinal and genitourinary tracts. C. auris, an emerging Candida strain, was first discovered in 2009 in Southeast Asia and is now present in 33 countries across six continents. The mortality rate of C. auris infection is high since it is resistant to almost all antifungals available, it can grow invasively and causes skin infections [7].
–
In the developing world, there are ∼1 million cases of cryptococcal diseases per year resulting in 675,000 deaths [8][9]. Cryoptococcus neoformans is an opportunistic fungal pathogen that causes meningitis in immunocompromised individuals. Often found in soils contaminated with bird feces, C. neoformans enters its host through the lungs via inhalation of spores. Some of the cryptococcal species are hypervirulent [10] and have drawn a considerable public attention due to their causative role in the cryptococcosis outbreak throughout the Pacific Northwest [11][12]. Only few antifungals are useful to treat cryptococcosis and drug resistant strains are emerging.
–
Aspergillus spp. are ubiquitous molds found widely in the environment as saprophytes and produce microscopic spores or conidia which, upon inhalation, cause invasive pulmonary disease. In immunocompromised patients having hematopoietic stem cell transplantation, solid organ transplantation, or chemotherapy, invasive aspergillosis remains the major cause of infection-related mortality [13][14]. Among several species of Aspergillus, A. fumigatus and A. flavus are common pathogens.
–
Dermatophytes are another group of keratinophilic pathogenic fungi that cause a variety of infections in humans and animals [15]. Some of these fungi include Trichophyton tonsurans (scalp ring-worm), T. equinum, and Microsporum gypseum (garderner’s ringworm). Emerging fungal diseases such as zygomycosis are life-threatening particularly during natural calamity (e.g. the 2004 tsunami, the 2008 Katrina and May 2011 Joplin tornado). Molecules with broad-spectrum antifungal activity are highly desirable to combat various fungal pathogens.
–
Because fungi are eukaryotes, the development of antifungal therapeutics that are nontoxic to humans is challenging due to the availability of relatively few targets. In the last twenty years, only one new class of antimycotic (β-glucan synthase inhibitor, the echinocandins) was introduced into clinical practice. Although this drug is an important addition, it has a number of limitations including ineffectiveness against Cryptococcus spp. and poor oral bioavailability [16]. Currently, the antifungal therapeutic options are limited, especially when compared to available antibacterial agents [2][17][18][19]. Among the five classes of antifungals, azoles, echiocandins, polyenes, allylamines, and pyrimidine derivatives, only three are used clinically: azoles, echiocandins, and polyenes. Azole drugs, such as fluconazole (FLU), inhibit ergosterol synthesis through inhibition of lanosterol 14-α-demethylase, impairing formation of the fungal cell membrane. Echocandins, such as caspofungin (CAS), block 1,3-β-glucan synthase and lead to depletion of glucan in the fungal cell wall. Polyenes, including amphotericin B (AMB), bind to ergosterol in the fungal cell membrane and change the cell membrane transition temperature, resulting in the leakage of ions and small organic molecules, and eventual cell death. Allylamines, such as amorolfin, affect ergosterol synthesis by the inhibition of squalene epoxidase. Pyrimidines, such as flucytosine (or 5-fluorocytosine), block nucleic acid synthesis, leading to the impediment of protein synthesis [20][21]. Although a new antifungal drug, isavuconazonium sulfate, belonging to azole family, has been developed in 2015 [22], no new ‘class’ of antifungal agent has been approved by the Food and Drug Administration (FDA) since 2006 [18][23][24].. Thus, the invention of new antifungal classes to overcome the increasing emergence of antifungal drug resistance is greatly needed.
–
Traditional antimycotics have drawbacks, including toxicity to human cells, a limited range of cellular targets, the development of antifungal resistance [3][9][25][26], and the failure to successfully control pathogenesis. To develop new antifungal agents based on drimane sesquiterpenes, we have investigated synthetic drimane terpenes, (-)-drimenol (1) and (+)-albicanol (2), along with six analogs, 3– 8 (Figure 1A), for their antifungal activities and identified (-)-1 as a potent broad-spectrum fungicidal agent. Moreover, we determined their mechanism of action through forward genetic screening of mutant libraries of C. albicans and baker’s yeast and found that (-)-1 affects Crk1 kinase-dependent gene products involved in protein secretion and vacuolar biogenesis in fungi.
–
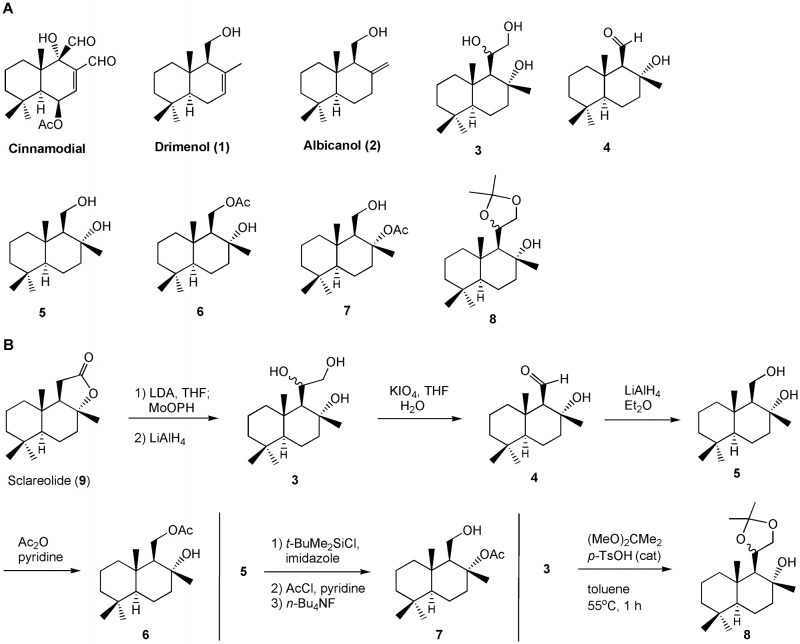 | FIGURE 1: Bioevaluation of drimane sesquiterpenoids for their antifungal activities (A) and their (3 – 8) synthesis (B). |
RESULTS
Several drimane sesquiterpenoids were synthesized in our laboratory during the total synthesis of (+)-chloropuupehenone, a natural product from marine sponges [27]. Based on the antimycobacterial activity of sesquiterpene natural product, cinnamodial, isolated from Warburgia salutaris (a species of tree in the Canellaceae family) [28], we anticipated that drimane sesquiterpenes and closely related compounds [29] could be effective antimycotics against C. albicans. Two other related antifungal compounds, muzigadial and warburganal, were also isolated from the Warburgia ugandensis plant [30]. Five representative drimane terpenes, 1– 5, along with their derivatives, 6– 8 (Figure 1A), were screened for their ability to inhibit C. albicans growth. It was assumed that additional hydroxyl group(s) or oxygen atoms in the molecule enhances water solubility and may improve bioactivity [31]. Molecules 3– 7 possess extra hydroxyl, aldehyde, or acetoxy functions and molecule 8 contains an acetonide moiety in the drimane structure.
–
Compounds 1– 5 were prepared by following previously reported methods [27]. Molecule 6 was made from a mono-acetylation of 5 with acetic anhydride and pyridine in dichloromethane (Figure 1B). Molecule 7 was obtained by a sequence of three reactions: (i) silylation of the primary alcohol with t-butyldimethylsilyl chloride and imidazole in dichloromethane; (ii) acetylation of the tertiary alcohol with acetyl chloride and pyridine; and (iii) removal of the silyl ether group with tetra-n-butylammonium fluoride in THF. Compound 8 was produced from the treatment of triol 3 with 2,2-dimethoxylpropane and a catalytic amount of p-toluenesulfonic acid in toluene. The experimental procedures are described in the Materials and Methods section.
–
Examination of the antifungal activities of these molecules along with the mechanistic study of the most active molecule may allow future improvement in bioactivity and reduction in toxicity. We used the C. albicans strain SC5314 for our initial screening of antifungal activities of drimane sesquiterpenoids 1– 8. The compounds were solubilized in dimethyl sulfoxide (DMSO), 10 mg/ml, as stock solutions and stored at −20°C. Prior to assays, stock solutions of compounds were diluted to 200 – 12.5 µg/ml in the growth media for yeast antifungal assays using the Clinical and Laboratory Standards Institute (CLSI) M38-A2 method [32]. Fortuitously, we found (-)-drimenol (1) and (+)-albicanol (2) along with compound 4, inhibit C. albicans SC5314 growth (Table 1). Among these three compounds, we identified 1 being more potent (with a MIC value ∼30 µg/ml) than other compounds (∼60 µg/ml); therefore 1 was used for further studies including mechanistic investigation.
–
TABLE 1. Antifungal activities of drimane sesquiterpenoids 1 – 8 against C. albicans SC5314. |
 |
–
Antifungal activities of drimenol against various pathogenic fungi
Since our initial assay with C. albicans confirmed the antifungal activities of 1, we extended the susceptibility assays to other pathogenic fungi including FLU resistant C. albicans, various species of Candida, Cryptococcus, Aspergillus, and a dermatophyte fungus. The CLSI broth dilution methods of M27-A3 for yeasts and M38-A2 for filamentous fungi [32] were used to determine the susceptibility. Molecule 4 was not further investigated due to the presence of an aldehyde function, which may react with biological molecules.
–
Briefly, yeast cells or conidia (for filamentous fungi) were suspended in RPMI-1640 medium to a final concentration of 105 cfu/ml and distributed in 96-well microplates to a total volume of 100 μl/well. Molecule 1 was added to the wells and two-fold serial dilutions were prepared. Du-plicates were used for each concentration and wells with or without DMSO served as controls. Plates were incubated without shaking at 37°C for 24 – 48 h for yeasts and 30°C for four days for filamentous fungi (Aspergillus and Trichophyton spp.). The MIC was defined as the lowest compound concentration at which no growth occurred, as determined visually and microscopically (inverted microscope). Representative images for the inhibition of fungi are shown in Supplementary Figure 1. Table 2 summarizes the MIC values of 1 and positive antifungal controls including FLU, posaconazole, and voriconazole, along with additional fungi such as C. glabrata [BG2], C. albicans [FLU resistant], C. auris, and T. equinum.
–
–
Results summarized in Supplementary Figure 1 show that 1 has a broad-spectrum fungicidal activity against various fungi including FLU resistant C. albicans and Cryptococcus spp. albeit, at a higher MIC concentration (50 μg/ml). However, for a dermatophyte fungus, the MIC value was lower (15 μg/ml). When compared to DMSO controls, fungi exposed to 1 showed an absence of growth. At higher concentration of 1 (100 μg/ml), C. albicans yeast cells lysed and released their cellular contents (Supplementary Figure 2, arrow). Consistent with this observation, 1 inhibited the germination of A. nidulans spores and appeared to cause swelling of germinating spores (Supplementary Figure 2, lower right).
–
–
To extend our antifungal screening with 1 against additional human pathogenic fungi, we have used the non-clinical and pre-clinical service program offered by the NIH NIAID supported fungus testing center at the University of Texas Health Sciences Center, San Antonio. The fungus testing center used the CLSI M38-A2 method [32] to determine the MIC of 1 after incubation for 24 – 72 hours with concentrations ranging from 0.125 – 64 μg/ml. Positive control antifungals (FLU, posaconazole and voriconazole) were also included in parallel. Results are summarized in Table 2. While the MIC for many fungi ranged from 4 – 64 μg/ml, for some fungi (Fusarium, Scedosporium, and Apophysomyces) it was above the highest concentration used (64 μg/ml). The antifungal susceptibility of these fungi to clinical antifungals (e.g. FLU) could be less.
–
Next, we determined the viability of fungal cells that were exposed to 1. Cells exposed to 50 μg/ml (MIC) for 24 h or at 100 μg/ml for 48 h were used. To determine the viability of treated fungal cells, small volumes (1 – 5 μg/ml) of mixed cell suspensions were removed from wells and spotted on YPD agar medium. The agar plates were incubated at 30°C for 24 h – 72 h and the growth of fungi was recorded. Growth of yeasts occurred within 24 h and of filamentous fungi within 48 – 72 h for control (without 1), but not for those treated with 1 suggesting that 1 acts as a fungicidal compound (data not shown).
–
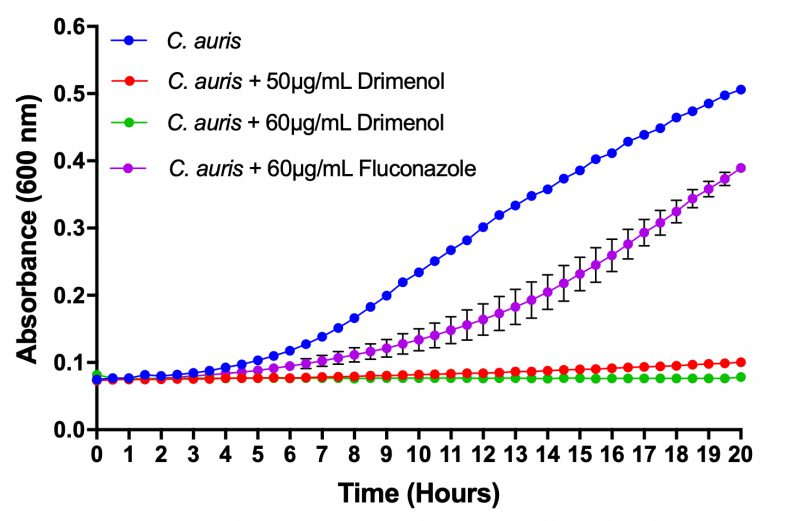
Drimenol acts better than FLU against C. auris growth
C. auris is an emerging multidrug resistant fungal pathogen that is known to cause nosocomial infections with “superbug”-like traits [33]. Recently, the Center for Disease Control and prevention has issued a clinical health emergency warning about this fungus. Since 1 showed a broad-spectrum fungicidal activity, we determined its effect against C. auris growth using a bioscreen-C growth monitoring system. C. auris was grown in the presence or absence of 1 in RPMI medium (CLSI method) [32] for 24 h at 37°C. Negative controls (solvent) and positive controls (FLU) were included in parallel. Results depicted in Figure 2 indicate that 1 inhibits C. auris growth completely at 60 μg/ml. In contrast, FLU at the same concentration (60 μg/ml) showed poor inhibition of growth. Thus, 1 could be useful as a broad-spectrum fungicidal compound.
–
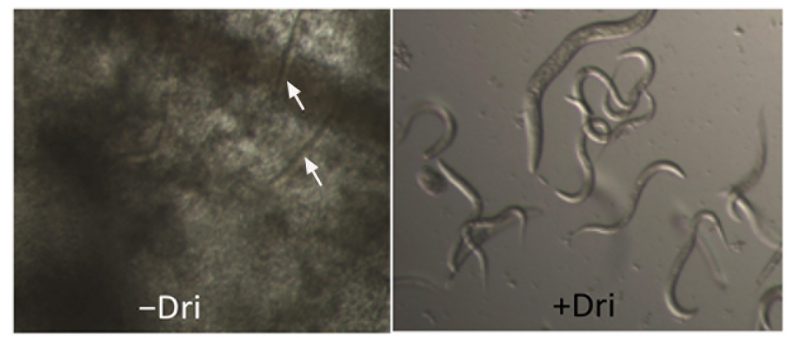
Drimenol (1) is tolerated by Caenorhabditis elegans and protects it from fungal-mediated death
Invertebrate animal models provide an inexpensive and powerful platform to test antifungal compounds for their efficacy and toxicity simultaneously. We evaluated 1 for its antifungal activity and tolerance in a C. elegans infection model of candidiasis as described previously [34]. Results shown in Figure 3 indicate that 1 can protect worms from C. albicans-mediated death and that the worms were not adversely affected by 1, as judged by their motility and viability following compound exposure. To test the toxicity of 1 against the nematode, they were incubated with different concentrations of 1 (100, 50, 25, 12, and 6 μg/ml) for 24 h in M9 medium. 1 affected the worms very little (∼6% death and ∼94% survived) at the maximum concentration used (100 μg/ml).
–
–
Mechanism of drimenol (1) antifungal activity
To understand the compound’s mechanisms of action (MOA), researchers have used pooled libraries of genome-wide barcoded mutant collections of Saccharomyces cerevisiae or C. albicans for drug-induced sensitivity assay or the haploinsufficiency (HIP) assay [35][36][37]. For example, if 1 can inactivate partially or completely its protein target in the heterozygous mutant pool, the resulting growth defect of that mutant(s) can be measured quantitatively by hybridization or sequencing the tagged unique barcodes. This approach will help narrow down the putative target(s). Similarly, a homozygous nonessential mutant library can be used as a complementary approach to the heterozygous essential mutant collection to verify the target pathway/genes of compounds. In this case, if the homozygous mutant of a gene is sensitive to the compound, then that gene may not be the drug target [35] as the homozygous mutant lacks the gene product. This implies that the compound may exert its effect via drug-induced synthetic lethality. Thus, by combining data from both heterozygote and homozygote screens one may determine the compound’s MOA.
–
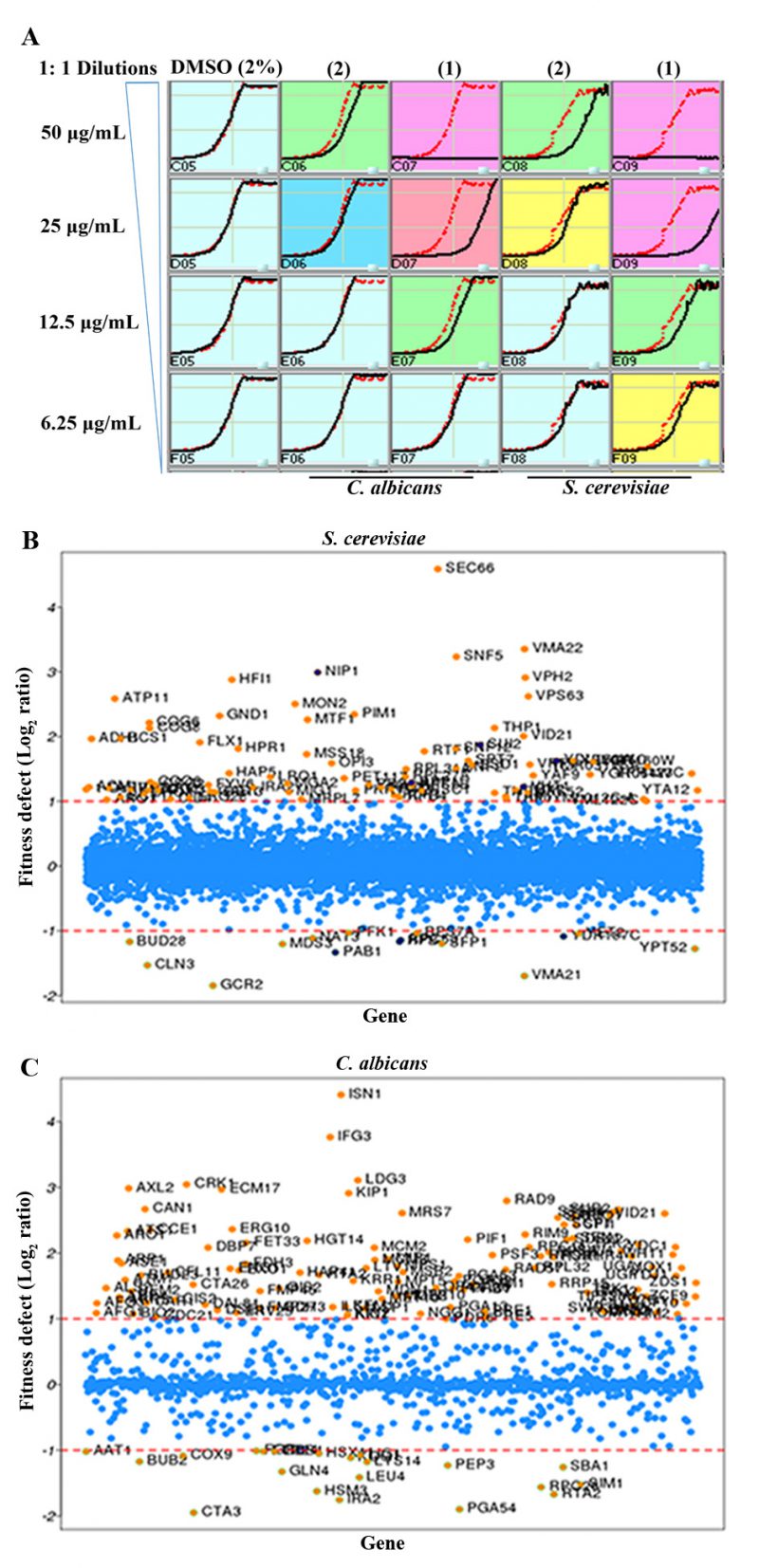
In this study, we used S. cerevisiae barcoded homozygous nonessential and heterozygous essential, and C. albicans barcoded heterozygous Tn mutant [37] libraries. Briefly, IC-50 of 1 for C. albicans and S. cerevisiae was determined under yeast growth (YPD) conditions (Figure 4A). Based on this assay results, IC-50 of 25 μg/ml for C. albicans and 15 μg/ml for S. cerevisiae was calculated for 1. Two different sub-MIC concentrations of 1 were selected for determining the mechanism of action against mutant libraries.
–
–
Next, pooled S. cerevisiae and C. albicans mutant collections were grown separately in the presence or absence of compounds (with DMSO) for 20 generations, barcodes from genomic DNA were amplified and relative strain abundances were quantified based on TAG microarray signals. The log2 ratio of tag signals between DMSO control and 1 exposed samples is presented in scatter plots as the “fitness defect” (Figures 4B and C). Mutants that were depleted from the growth pool due to 1 are indicated by circles. Mutants that were highly susceptible to 1 are shown with high log ratios (e.g. SEC66 in Figure 4B; highly depleted in the pool) and considered putative targets. Lists of S. cerevisiae and C. albicans mutants that are highly sensitive to 1 are listed in Supplementary Tables 1 and 2, respectively.
–
Our results of forward genetic screening from mutant libraries of C. albicans and S. cerevisiae with 1 indicate that it affects cellular activities involved in protein secretion, vacuolar functions, chromatin remodeling and cyclin dependent protein kinase (CDK)-associated functions (Figure 5A and Supplementary Tables 1 and 2). For example, SEC66, highly sensitive to 1 (log2>4.5, Figure 4B), is a component of Sec63 SECretary complex in S. cerevisiae, involved in protein targeting and import into the ER. Similarly, VMA22 is a vacuolar membrane ATPase required for vacuolar H+-ATPase function and localized to the yeast ER (Saccharomyces Genome Database).
–
–
The ISN1 (log2 4.4, Figure 4C) gene product is involved in inosine 5′-monophosphate 5′-nucleotidase activity in C. albicans (Candida Genome Database). This gene product is uncharacterized and it is present only in fungi and not in human or murine cells, suggesting that Isn1p a suitable antifungal drug target. IFG3 is a putative D-amino acid oxidase, which is uncharacterized, and CRK1 is a protein kinase of the Cdc2 subfamily involved in hyphal development and virulence in C. albicans[38]. The CRK1 ortholog in S. cerevisiae is SGV1, which is a part of the BUR2 kinase complex and plays a major role in transcriptional regulation.
–
Yeasts spot assay to validate drimenol (1) mechanism of action
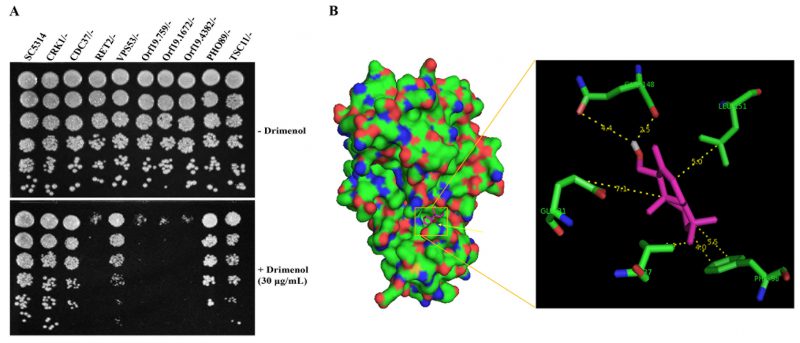
Based on the forward genetic library screening assay results (Figure 4) and the functions of putative targets inferred from the available literature, we selected a few heterozygous mutants of C. albicans that had high to medium positive log2 ratios (hypersensitive, CRK1 and its putative interacting partners proteins CDC37, Orf19.759, Orf19.1672 and Orf19.4382) and a few with negative log2 ratios (resistance, VPS53, TSC11& PHO89) to verify the genetic screening data. Agar medium containing a sub-MIC concentration of 1 (30 μg/ml) or solvent was used to spot test suspensions of various mutants (GRACE mutant collection) [39].
–
Results shown in Figure 5A confirm the findings of the forward genetic screening assays of C. albicans mutants. For example, Crk1 Kinase is predicted to associate with RET2, which is involved in retrograde vesicle transport for protein signaling and secretion (SEC66), and plays a role in protein translation with G1/S cell-cycle transition [40]. Products encoded by Ret2, orf19.759 (SEC21), orf19.1672 (COP1) and orf19.4382 (RET3) are uncharacterized and are likely targets of Crk1 kinase, which are defective in growth on agar medium containing 1 (Figure 5A). Molecule 1 induced hypersensitivity of these mutants, which represent candidate targets/pathways. The orf19.759 (SEC21 ortholog of S. cerevisiae) is uncharacterized in C. albicans. Sec21 is involved in the anterograde transport of vesicles from the endoplasmic reticulum (ER) to the Golgi, in the retrograde transport from the Golgi to the ER, COPI vesicle coat, and endosome localization [41]. RET2 is also uncharacterized in C. albicans and the ortholog is a subunit of the coatomer complex (COPI), which coats Golgi-derived transport vesicles, is involved in retrograde transport between the Golgi and the ER, and interacts with Crk1 kinase in the two-hybrid system [42]. Crk1 is known to play a role in regulating trafficking and secretion of effectors by interacting with the early endosome during Ustilago maydis (corn smut fungus) infection in corn plants [43][44].
–
Since Crk1 kinase may interact with multiple targets (Ret2, orf19.759, orf19.1672 and orf19.4382) and because Crk1 represents an important antifungal drug target [38], we performed computational molecular docking of 1 using the Cryptosporidium parvum Crk1 crystal structure (2QKR-A) [45] and the AutoDock Vina software [46]. Results shown in Figure 5B, suggest that 1 interacts with the N-terminal catalytic domain of C. albicans Crk1 (which has 61% similarity and 40% identity to the C. parvum Crk1). Particularly noteworthy from our computational docking studies is that 1 shows close interactions with Gly 31, Val 37, Gln 148, Leu 151, and Phe 98 amino acid residues. The docked structure shows that an available open space in Crk1 for incorporation of an additional functional group to the cyclohexane A ring of 1 (Figure 1), signifying a possible modification of 1 for future improvement of biological activity. Thus, this CDK member may comprise the target of 1; of note, this conserved gene is present in many of the tested pathogenic fungi (Table 2).
DISCUSSION
In our search for novel antifungal small molecules from our available synthetic terpenoids, we have identified two compounds, (-)-drimenol (1) and (+)-albicanol (2) (Figure 1), that show strong activity against C. albicans. Among these two compounds, 1 shows stronger bioactivity. It acts not only against C. albicans in a fungicidal manner but also against A. nidulans, FLU resistant strains of C. albicans, C. glabrata, C. krusei, Cryptococcus spp. and dermatophytes, suggesting that 1 is a broad-spectrum antifungal agent (Supplemenatary Figures 1, 2 and Table 2). At an increased concentration (100 μg/ml), 1 causes rupturing of the fungal cell wall/membrane, e.g. in C. albicans (Supplemenatary Figure 2) and Cryptococcus spp. (data not included). C. auris is an emerging and multidrug resistant strain that causes nosocomial infections and has been reported recently across the world [33]. Our bioscreen-based growth curve monitoring assay with 1 showed better activity than the clinically used antifungal drug FLU (Figure 2) indicating a potential use of 1 against C. auris and other drug-resistant fungal pathogens. Since molecule 1 is effective against the antifungal resistant C. auris, C. albicans and certain strains of C. neoformans (Table 2) and the MOA of 1 is different from FLU or other clinical antifungals, molecule 1 can be a useful additional antifungal agent with novel target(s). Molecule 1 synergizes FLU activity (FICI <0.5) in a checkerboard assay against C. albicans (data not shown) suggesting its potential utility in combinatorial antifungal therapy.
–
To understand the broad-spectrum antifungal potential, we evaluated 1 against various fungi that are pathogenic to humans and determined its mechanisms of action in C. albicans and S. cerevisiae. Based on our yeast mutant screening data and subsequent spot assay results, we found that 1 acts as a fungicidal compound by affecting cellular activities targeting protein trafficking between Golgi and ER, protein secretion (Sec system) and cell signaling, possibly through cell division related kinase 1, Crk1 (Figures 4B and C and 5A). Genetic methods have been used to determine the mechanism of antifungal compounds by drug-induced hypersensitivity assays [36][47]. Using similar approaches, we showed that 1-mediated inhibition of C. albicans heterozygous mutants of CDC37, Orf19.759, Orf19.1672 and Orf19.4382, the known or putative targets of Crk1 kinase, at sub-MIC concentration. Ac-cording to Nelson [48], the Cdc2 kinase (Crk1 kinase is a member of Cdc2 subfamily) plays a major role in regulating the retrograde membrane flow from the Golgi to the ER either alone or with another kinase kinase (e.g. MEK1) during mitosis. We speculate that molecule 1 may likely be disrupting the interaction of Crk1 with one or more of these gene products. In support of this observation, computational molecular docking of 1 with the crystal structure of a fungal (C. parvum) Crk1 kinase showed interactions of 1 with the key residues in the catalytic domain (N-terminal) of Crk1 (Figure 5B).
–
Molecule 1, a natural product, is present in liverworts and higher plants [49][50][51][52] and its antifungal activities against various pathogenic fungi have been reported previously though with higher MIC values. A recent study found that 1 had antifungal activity against Botrytis cinerea, a plant fungal pathogen, and the mechanisms appear to involve fungal membrane damage and reactive oxygen species (ROS) production in the germinating spores [52]. From our forward genetic screening and yeast spot assays, we identified Ret2 and related other gene products (orf19.759, orf19.1672, and orf19.4382) as hypersensitive to 1. Since Crk1 interacts with these SEC/COPI complexes [42][43][44] that traffic protein cargo between ER and Golgi, we surmise that molecule 1 by targeting these complexes may cause ROS production via ER-mediated unfolded protein response (UPR) or other pathways [54]. According to Robles-Kelly et al.[52], the ROS production in germinating spores due to drimenol treatment at 80 µg/ml was increased about 1-fold. However, we did not find the modulation of stress response (osmotic/oxidative) genes/pathways from our genetic screen data (Figure 4B and C, Supplementary Tables 1 and 2). The other possibility could be that physiological responses may vary between fungi (plant vs human pathogen), their growth conditions (media, spores vs yeast cells), and the concentrations of the drug used (20-25 µg/ml for yeasts vs 80 µg/ml for B. cinerea) [52]. Further studies are required to determine how molecule 1 affects these pathways.
–
Cinnamodial is a closely related compound belonging to the drimane sesquiterpenoid family with a potent antifungal activity [55], but its chemical structure (containing two aldehyde groups; Figure 1) and physiological properties are quite different from 1. For example, the antifungal activity of cinnamodial was shown to be abolished by amine compounds (likely due to a coupling reaction from the aldehyde functions of cinnamodial with the amino group of amine compounds) or when cinnamodial was used in YPD medium [4][56]. In contrast, molecule 1‘s bioactivity was not affected by amines or YPD medium (Figure 4A). Thus, the antifungal mechanisms of 1 could be different from cinnamodial. Since the synthetic route for 1 and its analogs are well established, improvements of its antifungal properties are possible through medicinal chemistry approaches and computational docking experiments.
–
In summary, we have synthesized a focused library of drimane sesquiterpenoid compounds and identified 1 as a broad-spectrum fungicidal compound against various human pathogenic fungi including C. albicans, C. auris, C. neoformans, Aspergillus, Blastomyces, Pneumocystis, and dermatophytes at concentrations of 8 – 64 µg/ml. By employing the libraries of bar-coded C. albicans and S. cerevisiae genome-wide mutants, the MOA of 1 was determined. Further evaluation of 1 in animal models of fungal diseases will help developing 1 as an antifungal agent.
MATERIALS AND METHODS
Synthesis of drimane molecules
(1R,2R,4aS,8aS)-2-Hydroxy-2,5,5,8a-tetramethyl-decahydro-naphthalene-1-carbaldehyde(4)
–
0.19 g (0.81 mmol) potassium periodate were added to a solution of 0.20 g (0.74 mmol) triol 3[27] in 10 ml THF and 2.5 ml water. The resulting mixture was stirred at 25°C for 4 hours, diluted with water (50 ml) and extracted three times with ethyl acetate (50 ml each). The combined extracts were washed with water and brine, dried (anhydrous Na2SO4), concentrated, and column chromatographed on silica gel using a mixture of hexane and ethyl acetate (20:1) as an eluent, yielding 0.16 g (91% yield) compound 4, whose spectral data is in agreement with that reported [27].
–
(1S,2R,4aS,8aS)-1-(Hydroxymethyl)-2,5,5,8a-tetramethyl-deca-hydronaphthalen-2-ol(5)
–
To a cold (0°C) solution of 1.0 g (4.2 mmol) aldehyde 4 in 80 ml of diethyl ether under argon, 80 mg (2.1 mmol) lithium aluminum hydride were added in portions. The resulting solution was stirred at 0°C for 30 minutes, diluted with aqueous NH4Cl, and extracted with diethyl ether three times (50 ml each). The combined extracts were washed with water and brine, dried (MgSO4), and concentrated, yielding 0.98 g (97% yield) diol 5, whose spectral data are in agreement with that reported [27].
–
[(1S,2R,4aS,8aS)-2-Hydroxy-2,5,5,8a-tetramethyl-decahydro-naphthalen-1-yl]methyl acetate(6)
–
To a cold (0°C) solution of 0.10 g (0.40 mmol) diol 5 in 2 ml dichloromethane and 0.32 g (4.0 mmol) pyridine under argon, 49 µl (0.48 mmol) of acetic anhydride were added, and the resulting solution was stirred at 0°C for 30 minutes and 25°C for 1 h. It was diluted with 30 ml of aqueous NH4OH, extracted twice with diethyl ether (30 ml each), and the combined extracts were washed with water and brine, dried (anhydrous Na2SO4), concentrated, and column chromatographed on silica gel using a gradient mixture of hexane and diethyl ether as eluents, yielding 90 mg (80% yield) of acetate 6. Mp. 64 – 67 °C; [α]D22= −8.2 (c = 0.55, CHCl3); 1H NMR (CDCl3; 400 MHz) α 4.35 (dd, J= 12, 4 Hz, 1 H), 4.24 (dd, J= 12, 4 Hz, 1 H), 2.05 (s, 3 H), 1.88 (dt, J= 12, 2 Hz, 1 H), 1.70 – 0.93 (a series of m, 11 H), 1.17 (s, 3 H), 0.88 (s, 3 H), 0.86 (s, 3 H), 0.80 (s, 3 H) ppm; 13C NMR (CDCl3; 100 MHz) α 171.4, 72.6, 62.6, 60.0, 55.7, 44.0, 41.7, 39.7, 38.1, 33.5, 33.2, 24.6, 21.6, 21.3, 20.3, 18.4, 15.8 ppm. MS (electrospray ionization), m/z 283.1 (M+H+). HRMS-ESI: m/z [M + H]+ calcd for C17H31O3+: 283.2268, found: 283.2273.
–
(1S,2R,4aS,8aS)-1-(Hydroxymethyl)-2,5,5,8a-tetramethyl-deca-hydronaphthalen-2-yl acetate(7)
–
Compound 7 was prepared by a sequence of three reactions: (i) silylation of the primary alcohol function of 5 with t-butyldimethylsilyl chloride; (ii) acetylation of the tertiary alcohol function with acetyl chloride and pyridine; and (iii) removal of the t-butyldimethylsilyl ether protecting group with tetra-n-butylammonium fluoride in THF.
–
To a solution of 9.5 mg (40 µmol) compound 5, 11 mg (150 µmol) imidazole, and 6 mg (49 µmol) 4-(dimethyl-amino)pyridine in 2 ml of dichloromethane under argon at 25°C, 14.3 mg (95 µmol) t-butyldimethylsilyl chloride were added, and the solution was stirred for 4 h. The reaction mixture was diluted with 10 ml of aqueous ammonium chloride and extracted with diethyl ether three times (10 ml each). The combined extracts were washed with water (10 ml) and brine (10 ml), dried (anhydrous Na2SO4), and concentrated, yielding 12.5 mg the mono-silylated product. This crude product was used in the subsequent step without purification. To a solution of the above mono-silylated product and 0.1 ml pyridine in 0.5 ml of dichloromethane under argon at 0°C, 10 µl (0.13 mmol) acetyl chloride were added. The reaction mixture was stirred at 25°C for 2 h, diluted with aqueous ammonium chloride (10 ml), and extracted three times with diethyl ether (10 ml each). The combined extracts were washed with brine, dried (anhydrous Na2SO4), and concentrated, getting the crude product, which was used in the following step without purification. The above crude product was dissolved in 1 ml of dried THF (distilled over sodium/benzophenone) and 0.3 ml (0.3 mmol) of tetra-n-butylammonium fluoride (1 M solution in THF) and stirred at 25°C under argon for 1 h. The reaction solution was diluted with 0.1 N ammonium hydroxide (10 ml) and extracted with diethyl ether three times (10 ml each). The combined extracts were washed with water (10 ml) and brine (10 ml), dried (anhydrous Na2SO4), concentrated, and column chromatographed on silica gel using a gradient mixture of hexane and diethyl ether, yielding 4.2 mg (38% overall yield from diol 5) of compound 7. Compound 7: Mp. 101 – 103 °C; [α]D22= +0.35 (c = 0.23, CHCl3); 1H NMR (CDCl3; 400 MHz) α 3.91 (dd, J= 12, 2 Hz, 1 H), 3.84 (dd, J= 12, 2 Hz, 1 H), 2.95 – 2.90 (m, 1 H), 1.98 (s, 3 H), 1.88 (dt, J= 12, 2 Hz, 1 H), 1.62 (s, 3 H), 1.70 – 0.88 (a series of m, 10 H), 0.94 (s, 3 H), 0.87 (s, 3 H), 0.82 (s, 3 H) ppm; 13C NMR (CDCl3; 100 MHz) α 169.9, 84.9, 63.8, 59.8, 55.8, 41.8, 39.5, 38.2, 36.1, 33.5, 33.2, 25.8, 22.8, 21.7, 18.3 (2 C), 16.1 ppm. MS (electrospray ionization), m/z 305.1 (M+Na+). HRMS-ESI: m/z [M + Na]+ calcd for C17H30NaO3+: 305.2087, found: 305.2082.
–
(1S,2R,4aS,8aS)-1-(2,2-Dimethyl-1,3-dioxolan-4-yl)-2,5,5,8a-tetramethyl-decahydronaphthalen-2-ol(8)
–
A solution of 18 mg (67 µmol) triol 3, 50 µl 2,2-dimethoxy-propane and 3 mg of anhydrous p-toluenesulfonic acid in 1 ml of toluene was stirred under argon at 55°C for 1 h. The solution was cooled to room temperature, neutralized with sodium bicarbonate (∼3 mg), diluted with 10 ml of water, and extracted with ethyl acetate three times (15 ml each). The combined extracts were washed with brine, dried (MgSO4), concentrated and column chromatographed on silica gel using a gradient mixture of hexane and diethyl ether as eluent, yielding 14 mg (71% yield) compound 8 as a mixture of two stereoisomers: (the major isomer was partially purified and reported) Mp. 114 – 117 °C; [α]D22= −25.1 (c = 1.0, CHCl3); 1H NMR (CDCl3; 400 MHz) α 4.96 (s, 1 H, OH), 4.24 – 4.20 (m, 2 H), 3.59 (td, J= 8, 4 Hz, 1 H), 1.84 (dt, J= 12, 2 Hz, 1 H), 1.70 – 0.83 (a series of m, 11 H), 1.45 (s, 3 H), 1.41 (s, 6 H, 2 CH3), 0.97 (s, 3 H), 0.90 (s, 3 H), 0.83 (s, 3 H) ppm; 13C NMR (CDCl3; 100 MHz) α 107.4, 73.5, 72.8, 62.2, 55.7, 42.8, 41.5, 40.4, 37.3, 33.6, 33.3, 26.5, 26.2, 25.8, 21.7, 19.7, 18.4 (2 C), 16.1 ppm. MS (electrospray ionization), m/z 333.1 (M+Na+). HRMS-ESI: m/z [M + Na]+ calcd for C19H34NaO3+: 333.2406, found: 333.2411.
–
Determination of the antifungal activity of the synthetic compounds
Synthetic pure drimenol or albicanol was dissolved in DMSO (10 mg/ml as stock solution) and used for determining their antifungal activities (minimum inhibitory concentration, MIC) against various fungi according to the microdilution assay of CLSI [32]. The CLSI broth dilution methods M27-A3 for yeasts and M38-A for filamentous fungi were used to determine the susceptibility. Since our initial assay with C. albicans confirmed the antifungal activity of drimenol and albicanol, we extended the susceptibility assay to other pathogenic fungi including FLU resistant C. albicans, various species of Candida, Cryptococcus, Aspergillus and a dermatophyte fungus [57] (strains were generously provided by Dr. Ted C. White at The University of Missouri Kansas City (UMKC). C. auris[58] was obtained from Dr. Baha Abdalhamid at The University of Nebraska Medical Center, Omaha NE). Briefly, yeast cells or conidia (for filamentous fungi) were suspended in RPMI-1640 medium to a final concentration of 105 cfu/ml and distributed in 96-well microplates to a total volume of 100 μl/well. Drimenol or albicanol was added into the wells and a two-fold serial dilution was made. Duplicates were used for each concentration and wells with or without DMSO served as controls. Plates were incubated without shaking at 37°C for 24 – 48h for yeasts and 30°C for four days for filamentous fungi (Aspergillus spp. and Trichophyton spp.). The MIC was defined as the lowest compound concentration at which no growth occurred, as determined visually and microscopically (inverted microscope).
–
Determination of C. auris growth inhibition by drimenol
The effect of drimenol on the growth of C. auris was determined by the Bioscreen-C real time growth monitoring system (Oy Growth Curves Ab Ltd, Finland) as described earlier [59]. The antifungal resistant C. auris strain [58] (South Asian Clade) [60][61] was used. Briefly, 200 µl of RPMI medium containing exponentially growing C. auris yeast cells (each at 0.07 OD600) were added into the honeycomb wells with or without compound (control) and growth rates were measured for 20 hours at 37°C. Compound treatment was done with two different concentrations for drimenol (50 and 60 µg/ml). The absorbance was measured at 600 nm in 30 min intervals for 24 h at 37°C with shaking for 10 s before each read. A solvent negative control (DMSO) and a FLU (60 µg/ml; antifungal drug) positive control were included in the study. The experiments were repeated at least two times with three technical replicates.
–
Caenorhabditis elegans (nematode) host model for the C. albicans infection assay
The nematode model of candidiasis was used as described before [34] to determine the antifungal efficacy of (1). Briefly, larvae (L2) were fed on a C. albicans yeast lawn on YPD agar plate. After collecting the larvae and washing off yeast cells with PBS buffer, an aliquot of larvae was mixed with buffered RPMI medium and distributed to microtiter wells. Drimenol (1) (50 μg/ml) or DMSO (solvent control) were added to microtiter wells equally containing the worms. As a positive antifungal control, amphotericin B (1 μg/ml) was included. Triplicate wells, about 20-30 nematodes/well, were used. The assay plate was placed in a plastic box lined with moisture paper and incubated at 30°C for 2-3 days. Nematodes were monitored under an inverted microscope and recorded with a digital camera connected to a microscope.
–
Yeast spot assay
Yeast Peptone Dextrose (YPD) agar containing a sub-MIC concentration of drimenol (30 µg/ml) or an equal volume of DMSO was used to spot test the C. albicans heterozygous mutants (GRACE library [39]). Yeast suspensions of various mutants and the wild type C. albicans were used. 5 μl of a four-fold serially diluted suspension were spotted on the agar plates and incubated at 30°C for yeast growth for 24 h, and photographed. Experiments were repeated at least three times and a representative result was shown.
–
Genome-wide fitness assay
The Saccharomyces yeast deletion collection was comprised of approximately 5,900 individually bar-coded heterozygous diploid strains (HIP [haploinsufficiency profiling]) and ∼4,800 homozygous diploid strains (HOP [homozygous deletion profiling]). Pools of approximately equal strain abundance were generated by robotically pinning (S and P Robotics, Ontario, Canada) each strain (from frozen stocks) onto YPD agar plates as arrays of 384 strains/plate [35][62][63]. After two days of growth at 30°C, colonies were collected from plates by flooding with YPD, and cells were adjusted to an optical density at 600 nm (OD600) of 2. The fitness of each strain in each experimental pool was assessed as described previously [35]. The dose that resulted in 15% growth inhibition in S. cerevisiae BY4743 (the parent strain of the yeast deletion collection) was determined by analyzing dose response over the course of 16 h of growth at 30°C. Screens of the homozygous deletion collection were performed over five generations of growth and screens of the heterozygous deletion collection were collected after 20 generations of growth. Cells were processed as described previously [35]. Genomic DNA was extracted from each sample and subjected to PCR to amplify the unique bar code identifiers. The abundance of each bar code was determined by quantifying the microarray signal as previously described [35]. C. albicans pooled screens used the tn-transposon collection [37]. Growth assays were performed in duplicates and samples were recovered after 20 generations of growth. Genomic DNA extraction, tag amplification, and hybridization were performed as described above.
REFERENCES
- Pfaller MA, Diekema DJ (2010). Epidemiology of invasive mycoses in North America. Crit Rev Microbiol 36(1): 1-53. 10.3109/10408410903241444
- Perlin DS, Rautemaa-Richardson R, Alastruey-Izquierdo A (2017). The global problem of antifungal resistance: prevalence, mechanisms, and management. Lancet Infect Dis 17(12): e383-e392. 10.1016/S1473-3099(17)30316-X
- Pfaller MA, Diekema DJ (2007). Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev 20(1): 133-163. 10.1128/CMR.00029-06
- Denning DW, Kneale M, Sobel JD, Rautemaa-Richardson R (2018). Global burden of recurrent vulvovaginal candidiasis: a systematic review. Lancet Infect Dis 18(11): e339-e347. 10.1016/S1473-3099(18)30103-8.
- Pappas PG (2006). Invasive candidiasis. Infect Dis Clin North Am 20(3): 485-506. S0891-5520(06)00062-6 [pii] 10.1016/j.idc.2006.07.004
- Pfaller MA, Diekema DJ, Rinaldi MG, Barnes R, Hu B, Veselov AV, Tiraboschi N, Nagy E, Gibbs DL (2005). Results from the ARTEMIS DISK Global Antifungal Surveillance Study: a 6.5-year analysis of susceptibilities of Candida and other yeast species to fluconazole and voriconazole by standardized disk diffusion testing. J Clin Microbiol 43(12): 5848-5859. 10.1128/JCM.43.12.5848-5859.2005
- de Jong AW, Hagen F (2019). Attack, Defend and Persist: How the Fungal Pathogen Candida auris was Able to Emerge Globally in Healthcare Environments. Mycopathologia 184(3): 353-365. 10.1007/s11046-019-00351-w
- Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM (2009). Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS 23(4): 525-530. 10.1097/QAD.0b013e328322ffac
- Denning DW, Hope WW (2010). Therapy for fungal diseases: opportunities and priorities. Trends Microbiol 18(5): 195-204. 10.1016/j.tim.2010.02.004
- Byrnes EJ, 3rd, Li W, Lewit Y, Ma H, Voelz K, Ren P, Carter DA, Chaturvedi V, Bildfell RJ, May RC, Heitman J (2010). Emergence and pathogenicity of highly virulent Cryptococcus gattii genotypes in the northwest United States. PLoS Pathog 6(4): e1000850. 10.1371/journal.ppat.1000850
- Kidd SE, Hagen F, Tscharke RL, Huynh M, Bartlett KH, Fyfe M, Macdougall L, Boekhout T, Kwon-Chung KJ, Meyer W (2004). A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada). Proc Natl Acad Sci U S A 101(49): 17258-17263. 0402981101 [pii] 10.1073/pnas.0402981101
- NPR-News (2010). Fungal disease spreads through Pacific Northwest. Available at : http://www.npr.org/templates/story/story.php?storyId=126198896. [Accessed 10.08.2019]
- Alangaden GJ (2011). Nosocomial fungal infections: epidemiology, infection control, and prevention. Infect Dis Clin North Am 25(1): 201-225. 10.1016/j.idc.2010.11.003
- Neofytos D, Horn D, Anaissie E, Steinbach W, Olyaei A, Fishman J, Pfaller M, Chang C, Webster K, Marr K (2009). Epidemiology and outcome of invasive fungal infection in adult hematopoietic stem cell transplant recipients: analysis of Multicenter Prospective Antifungal Therapy (PATH) Alliance registry. Clin Infect Dis 48(3): 265-273. 10.1086/595846
- Achterman RR, Smith AR, Oliver BG, White TC (2011). Sequenced dermatophyte strains: growth rate, conidiation, drug susceptibilities, and virulence in an invertebrate model. Fungal Genet Biol 48(3): 335-341. 10.1016/j.fgb.2010.11.010
- Butler MS (2008). Natural products to drugs: natural product-derived compounds in clinical trials. Nat Prod Rep 25(3): 475-516. 10.1039/b514294f
- Fuentefria AM, Pippi B, Dalla Lana DF, Donato KK, de Andrade SF (2018). Antifungals discovery: an insight into new strategies to combat antifungal resistance. Lett Appl Microbiol 66(1): 2-13. 10.1111/lam.12820
- Perfect JR (2017). The antifungal pipeline: a reality check. Nat Rev Drug Discov 16(9): 603-616. 10.1038/nrd.2017.46
- Nami S, Aghebati-Maleki A, Morovati H, Aghebati-Maleki L (2019). Current antifungal drugs and immunotherapeutic approaches as promising strategies to treatment of fungal diseases. Biomed Pharmacother 110: 857-868. 10.1016/j.biopha.2018.12.009
- Nett JE, Andes DR (2016). Antifungal Agents: Spectrum of Activity, Pharmacology, and Clinical Indications. Infect Dis Clin North Am 30(1): 51-83. 10.1016/j.idc.2015.10.012
- Waldorf AR, Polak A (1983). Mechanisms of action of 5-fluorocytosine. Antimicrob Agents Chemother 23(1): 79-85. 10.1128/aac.23.1.79
- Kovanda LL, Maher R, Hope WW (2016). Isavuconazonium sulfate: a new agent for the treatment of invasive aspergillosis and invasive mucormycosis. Expert Rev Clin Pharmacol 9(7): 887-897. 10.1080/17512433.2016.1185361
- Denning DW (2002). Echinocandins: a new class of antifungal. J Antimicrob Chemother 49(6): 889-891. 10.1093/jac/dkf045
- Roemer T, Krysan DJ (2014). Antifungal drug development: challenges, unmet clinical needs, and new approaches. Cold Spring Harb Perspect Med 4(5): a019703. 10.1101/cshperspect.a019703
- Marie C, White TC (2009). Genetic Basis of Antifungal Drug Resistance. Curr Fungal Infect Rep 3(3): 163-169. 10.1007/s12281-009-0021-y
- Sanglard D, Coste A, Ferrari S (2009). Antifungal drug resistance mechanisms in fungal pathogens from the perspective of transcriptional gene regulation. FEMS Yeast Res 9(7): 1029-1050. 10.1111/j.1567-1364.2009.00578.x
- Hua DH, Huang X, Chen Y, Battina SK, Tamura M, Noh SK, Koo SI, Namatame I, Tomoda H, Perchellet EM, Perchellet JP (2004). Total syntheses of (+)-chloropuupehenone and (+)-chloropuupehenol and their analogues and evaluation of their bioactivities. J Org Chem 69(18): 6065-6078. 10.1021/jo0491399
- Madikane VE, Bhakta S, Russell AJ, Campbell WE, Claridge TD, Elisha BG, Davies SG, Smith P, Sim E (2007). Inhibition of mycobacterial arylamine N-acetyltransferase contributes to anti-mycobacterial activity of Warburgia salutaris. Bioorg Med Chem 15(10): 3579-3586. 10.1016/j.bmc.2007.02.011
- Cortes M, Delgado V, Saitz C, Armstrong V (2011). Drimenol: A versatile synthon for compounds with trans-drimane skeleton. Nat Prod Commun 6(4): 477-490. 10.1177/1934578×1100600408
- Kubo I, Miura I, Pettei MJ, Lee YW, Pilkiewicz F, Nakanishi K (1977). Muzigadial and warburganal, potent antifungal antiyeast, and african army worm antifeedant agents. Tetrahedron Letters 52: 4553-4556. 10.1016/s0040-4039(01)83566-8
- Liu Y, Liu Z, Cao X, Liu X, He H, Yang Y (2011). Design and synthesis of pyridine-substituted itraconazole analogues with improved antifungal activities, water solubility and bioavailability. Bioorg Med Chem Lett 21(16): 4779-4783. 10.1016/j.bmcl.2011.06.062
- CLSI (2008). Reference method for broth dilution antifungal susceptibility testing of yeasts and filamentous fungi; Approved Standard-Third Edition M27-A3 & M38-A4. Clinical and Laboratory Standards Institute, Wayne, Pa.
- Kean R, Ramage G (2019). Combined Antifungal Resistance and Biofilm Tolerance: the Global Threat of Candida auris. mSphere 4(4). 10.1128/mSphere.00458-19
- Vediyappan G, Dumontet V, Pelissier F, d’Enfert C (2013). Gymnemic acids inhibit hyphal growth and virulence in Candida albicans. PLoS One 8(9): e74189. 10.1371/journal.pone.0074189
- Pierce SE, Davis RW, Nislow C, Giaever G (2007). Genome-wide analysis of barcoded Saccharomyces cerevisiae gene-deletion mutants in pooled cultures. Nat Protoc 2(11): 2958-2974. 10.1038/nprot.2007.427
- Xu D, Jiang B, Ketela T, Lemieux S, Veillette K, Martel N, Davison J, Sillaots S, Trosok S, Bachewich C, Bussey H, Youngman P, Roemer T (2007). Genome-wide fitness test and mechanism-of-action studies of inhibitory compounds in Candida albicans. PLoS Pathog 3(6): e92. 10.1371/journal.ppat.0030092
- Oh J, Fung E, Schlecht U, Davis RW, Giaever G, St Onge RP, Deutschbauer A, Nislow C (2010). Gene annotation and drug target discovery in Candida albicans with a tagged transposon mutant collection. PLoS Pathog 6(10): e1001140. 10.1371/journal.ppat.1001140
- Chen J, Zhou S, Wang Q, Chen X, Pan T, Liu H (2000). Crk1, a novel Cdc2-related protein kinase, is required for hyphal development and virulence in Candida albicans. Mol Cell Biol 20(23): 8696-8708. 10.1128/mcb.20.23.8696-8708.2000
- Roemer T, Jiang B, Davison J, Ketela T, Veillette K, Breton A, Tandia F, Linteau A, Sillaots S, Marta C, Martel N, Veronneau S, Lemieux S, Kauffman S, Becker J, Storms R, Boone C, Bussey H (2003). Large-scale essential gene identification in Candida albicans and applications to antifungal drug discovery. Mol Microbiol 50(1): 167-181. 10.1046/j.1365-2958.2003.03697.x
- An T, Liu Y, Gourguechon S, Wang CC, Li Z (2018). CDK Phosphorylation of Translation Initiation Factors Couples Protein Translation with Cell-Cycle Transition. Cell Rep 25(11): 3204-3214 e3205. 10.1016/j.celrep.2018.11.063
- CGD (2019). Candida Genome Database C. albicans SEC21/C1_04830W. Available at http://www.candidagenome.org/cgi-bin/locus.pl?locus=Sec21&organism=C_albicans_SC5314. [Accessed 06.10.2019]
- CGD (2019). Candida Genome Database C. albicans RET2/C5_02300C. Available at http://www.candidagenome.org/cgi-bin/locus.pl?locus=ret2&organism=C_albicans_SC5314. [Accessed 06.10.2019]
- Higuchi Y (2015). Initial fungal effector production is mediated by early endosome motility. Commun Integr Biol 8(2): e1025187. 10.1080/19420889.2015.1025187
- Bielska E, Higuchi Y, Schuster M, Steinberg N, Kilaru S, Talbot NJ, Steinberg G (2014). Long-distance endosome trafficking drives fungal effector production during plant infection. Nat Commun 5: 5097. 10.1038/ncomms6097
- Wernimont AK, Dong A, Lew J, Lin YH, Hassanali A, Arrowsmith CH, Edwards AM, Weigelt J, Sundstrom M, Bochkarev A, Hui R, Artz JD (2007). Cryptosporidium parvum cyclin-dependent kinase cgd5_2510 with indirubin 3′-monoxime bound. Structural Genomics Consortium (SGC). Available at: https://wwwrcsborg/structure/2qkr. [Accessed 05.09.2019]
- Trott O, Olson AJ (2010). AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31(2): 455-461. 10.1002/jcc.21334
- Giaever G, Shoemaker DD, Jones TW, Liang H, Winzeler EA, Astromoff A, Davis RW (1999). Genomic profiling of drug sensitivities via induced haploinsufficiency. Nat Genet 21(3): 278-283. 10.1038/6791
- Nelson WJ (2000). W(h)ither the Golgi during mitosis? J Cell Biol 149(2): 243-248. 10.1083/jcb.149.2.243
- Jansen BJ, de Groot A (2004). Occurrence, biological activity and synthesis of drimane sesquiterpenoids. Nat Prod Rep 21(4): 449-477. 10.1039/b311170a
- Toyota M, Ooiso Y, Kusuyama T, Asakawa Y (1994). Drimane-type sesquiterpenoids from the liverwort Diplophyllum serrulatum.Phytochemistry 35(5): 1263-1265. 10.1016/s0031-9422(00)94833-7
- Derita M, Montenegro I, Garibotto F, Enriz RD, Fritis MC, Zacchino SA (2013). Structural requirements for the antifungal activities of natural drimane sesquiterpenes and analogues, supported by conformational and electronic studies. Molecules 18(2): 2029-2051. 10.3390/molecules18022029
- Robles-Kelly C, Rubio J, Thomas M, Sedan C, Martinez R, Olea AF, Carrasco H, Taborga L, Silva-Moreno E (2017). Effect of drimenol and synthetic derivatives on growth and germination of Botrytis cinerea: Evaluation of possible mechanism of action. Pestic Biochem Physiol 141:50-56. 10.1016/j.pestbp.2016.11.006
- Hu H, Gourguechon S, Wang CC, Li Z (2016). The G1 Cyclin-dependent Kinase CRK1 in Trypanosoma brucei Regulates Anterograde Protein Transport by Phosphorylating the COPII Subunit Sec31. J Biol Chem 291(30): 15527-15539. 10.1074/jbc.M116.715185
- Malhotra JD, Miao H, Zhang K, Wolfson A, Pennathur S, Pipe SW, Kaufman RJ (2008). Antioxidants reduce endoplasmic reticulum stress and improve protein secretion. Proc Natl Acad Sci U S A 105(47): 18525-18530. 10.1073/pnas.0809677105
- Kubo I, Fujita K, Lee SH (2001). Antifungal mechanism of polygodial. J Agric Food Chem 49(3): 1607-1611. 10.1021/jf000136g
- Fujita K, Kubo I (2005). Multifunctional action of antifungal polygodial against Saccharomyces cerevisiae: involvement of pyrrole formation on cell surface in antifungal action. Bioorg Med Chem 13(24): 6742-6747. 10.1016/j.bmc.2005.07.023
- Achterman RR, White TC (2013). Dermatophytes. Curr Biol 23(13): R551-552. 10.1016/j.cub.2013.03.026
- Abdalhamid B, Almaghrabi R, Althawadi S, Omrani A (2018). First report of Candida auris infections from Saudi Arabia. J Infect Public Health 11(4): 598-599. 10.1016/j.jiph.2018.05.010
- Veerapandian R, Vediyappan G (2019). Gymnemic acids inhibit adhesive nanofibrillar mediated Streptococcus gordonii-Candida albicans mono-species and dual-species biofilms. Front Microbiol 10: 2328. 10.3389/fmicb.2019.02328
- Emara M, Ahmad S, Khan Z, Joseph L, Al-Obaid I, Purohit P, Bafna R (2015). Candida auris candidemia in Kuwait, 2014. Emerg Infect Dis 21(6): 1091-1092. 10.3201/eid2106.150270
- Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, Govender NP, Colombo AL, Calvo B, Cuomo CA, Desjardins CA, Berkow EL, Castanheira M, Magobo RE, Jabeen K, Asghar RJ, Meis JF, Jackson B, Chiller T, Litvintseva AP (2017). Simultaneous Emergence of Multidrug-Resistant Candida auris on 3 Continents Confirmed by Whole-Genome Sequencing and Epidemiological Analyses. Clin Infect Dis 64(2): 134-140. 10.1093/cid/ciw691
- Mor V, Rella A, Farnoud AM, Singh A, Munshi M, Bryan A, Naseem S, Konopka JB, Ojima I, Bullesbach E, Ashbaugh A, Linke MJ, Cushion M, Collins M, Ananthula HK, Sallans L, Desai PB, Wiederhold NP, Fothergill AW, Kirkpatrick WR, Patterson T, Wong LH, Sinha S, Giaever G, Nislow C, Flaherty P, Pan X, Cesar GV, de Melo Tavares P, Frases S, et al. (2015). Identification of a New Class of Antifungals Targeting the Synthesis of Fungal Sphingolipids. MBio 6(3): e00647. 10.1128/mBio.00647-15e00647-15
- Proctor M, Urbanus ML, Fung EL, Jaramillo DF, Davis RW, Nislow C, Giaever G (2011). The automated cell: compound and environment screening system (ACCESS) for chemogenomic screening. Methods Mol Biol 759: 239-269. 10.1007/978-1-61779-173-4_15
–
SUPPLEMENTAL INFORMATION
![]() Download Supplemental Information
Download Supplemental Information
ACKNOWLEDGMENTS
We thank Ted C. White, University of Missouri Kansas City (UMKC), Division of Cell Biology and Biophysics, Kansas City for various species of candida including fluconazole-resistant strains, Cryptococcus, Aspergillus and dermatophytes. Our sincere thanks to Terry Roemer, Merck Sharp and Dohme Corp, NJ for providing C. albicans GRACE and DBC mutant libraries. C. auris was obtained from Baha Abdalhamid, University of Nebraska Medical Center, Omaha NE. Kansas State Uni-versity has utilized the non-clinical and pre-clinical ser-vice program offered by the National Institute of Allergy and Infectious Diseases at the Fungus Testing Laboratory, UTHSC San Antonio. Our sincere thanks to Nathan Wiederhold, Hoja Patterson, and April Torres for their assistance in testing drimenol for antifungal activity through the NIH service program. We thank Johnson Cancer Research Center, KSU for Innovative Research Award and Kansas Idea Network of Biomedical Research Excellence (K-INBRE) for CORE Facility support award to GV. CN is a CRC Tier 1 Chair in Translational Genomics and support for GG and CN is provided by the CIHR. Re-search reported in this publication was, in part, support-ed by the National Institute of General Medical Sciences of the National Institutes of Health under award number NIH R01GM128659 (to DHH). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This material was based upon work in part sup-ported by the National Science Foundation under 1826982 (to DHH) for the purchase of a 400-MHz NMR spectrometer.
COPYRIGHT
© 2020

Broad-spectrum antifungal activities and mechanism of drimane sesquiterpenoids by Edouarzin et al. is licensed under a Creative Commons Attribution 4.0 International License.