Research Articles:
Microbial Cell, Vol. 3, No. 1, pp. 29 - 45; doi: 10.15698/mic2016.01.473
Global translational impacts of the loss of the tRNA modification t6A in yeast
1 Department of Microbiology and Cell Science, University of Florida, Gainesville, FL 32611, USA.
2 Genetics and Genomics Graduate Program, University of Florida, Gainesville, FL 32610, USA.
3 University of Florida Genetics Institute, University of Florida, Gainesville, FL 32610, USA.
4 Institut de Biologie Intégrative de la Cellule (I2BC), CEA, CNRS, Université Paris-Sud, Bâtiment 400, 91400 Orsay, France.
5 Centro de Regulación del Genoma. Facultad de Ciencias – Universidad de Chile, Santiago, Chile.
6 Laboratory of Molecular Biology, Francis Crick Avenue, Cambridge CB2 0QH, United Kingdom.
Keywords: t6A, tRNA, ribosome profiling, translation, modified nucleosides.
Abbreviations:
AGEs - advanced glycation end-products,
ASL - anticodon stem loop,
CO - codon occupancy,
RRT - ribosome residence time,
RPFs - ribosome-protected fragments,
tRNA - transfer RNA,
UPR - unfolded protein response.
Received originally: 10/11/2015 Received in revised form: 19/11/2015
Accepted: 19/11/2015
Published: 18/12/2015
Correspondence:
Olivier Namy, Institut de Biologie Intégrative de la Cellule (I2BC), CEA, CNRS, Université Paris-Sud, Bâtiment 400; 91400 Orsay, France olivier.namy@igmors.u-psud.fr
Valérie de Crécy-Lagard, Department of Microbiology and Cell Science, University of Florida, P.O. Box 110700; Gainesville, FL 32611-0700, USA vcrecy@ufl.edu
Conflict of interest statement: The authors declare no conflict of interest.
Please cite this article as: Patrick C. Thiaville, Rachel Legendre, Diego Rojas-Benítez, Agnès Baudin-Baillieu, Isabelle Hatin, Guilhem Chalancon, Alvaro Glavic, Olivier Namy, Valérie de Crécy-Lagard (2015). Global translational impacts of the loss of the tRNA modification t6A in yeast. Microbial Cell 3(1): 29-45.
Abstract
The universal tRNA modification t6A is found at position 37 of nearly all tRNAs decoding ANN codons. The absence of t6A37 leads to severe growth defects in baker’s yeast, phenotypes similar to those caused by defects in mcm5s2U34 synthesis. Mutants in mcm5s2U34 can be suppressed by overexpression of tRNALysUUU, but we show t6A phenotypes could not be suppressed by expressing any individual ANN decoding tRNA, and t6A and mcm5s2U are not determinants for each other’s formation. Our results suggest that t6A deficiency, like mcm5s2U deficiency, leads to protein folding defects, and show that the absence of t6A led to stress sensitivities (heat, ethanol, salt) and sensitivity to TOR pathway inhibitors. Additionally, L-homoserine suppressed the slow growth phenotype seen in t6A-deficient strains, and proteins aggregates and Advanced Glycation End-products (AGEs) were increased in the mutants. The global consequences on translation caused by t6A absence were examined by ribosome profiling. Interestingly, the absence of t6A did not lead to global translation defects, but did increase translation initiation at upstream non-AUG codons and increased frame-shifting in specific genes. Analysis of codon occupancy rates suggests that one of the major roles of t6A is to homogenize the process of elongation by slowing the elongation rate at codons decoded by high abundance tRNAs and I34:C3 pairs while increasing the elongation rate of rare tRNAs and G34:U3 pairs. This work reveals that the consequences of t6A absence are complex and multilayered and has set the stage to elucidate the molecular basis of the observed phenotypes.
INTRODUCTION
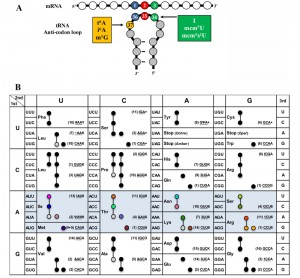
Modifications of the anticodon stem loop (ASL) of transfer RNA (tRNA) are critical for translational speed and accuracy. As the genetic code is degenerate, most tRNAs decode several codons [1]. Nucleoside modifications ensure that the decoding process is stringent enough to discriminate between closely-related codons and yet relaxed enough to allow decoding of more than one codon [2][3]. Different organisms use distinct but convergent strategies to optimize speed and accuracy of decoding by modifying specific tRNAs, predominantly at position 34 (the wobble base) and at position 37 (the dangling base) of the ASL (Figure 1A) [2][3]. Modifications at position 34, such as ncm5U34 (5-carbamoylmethyluridine) or I34 (inosine) can expand decoding capacity, thereby allowing one tRNA to decode three synonymous codons [2][3][4]. Likewise, most modifications at position 37 are critical for decoding, and their role can be complex. For example, i6A37 (N6-isopentenyladenosine) promotes decoding activity and increases fidelity of tRNACysGCA at its cognate codon, but also increases the misreading rate of tRNATyrGUA at the near-cognate UGC codon, which makes the effects of this modification on protein expression difficult to disentangle [5]. In fact, the exact in vivo contributions of many ASL modifications to translational robustness are still poorly understood [6].
Codon usage bias allows for fine-tuning of translation by ASL modifications. Codon choice (which codon in a synonymous set is used to encode a given amino acid) affects gene expression levels, protein production, accuracy, protein folding [7][8][9][10], and can even be used to predict gene function [11]. Codon usage bias is not only driven by neutral processes such as mutation biases or GC%, but is also molded by selection [9][12]. Although translation speed is a strong driving force of codon usage [9], the avoidance of codons with higher propensity for protein synthesis errors leading to misfolding is also an important factor in codon selection [13][14]. ASL modifications play key roles in both these processes [6][7]. In addition, systems level approaches integrating proteomics, codon usage, and modification profiling [15] have recently shown that tRNA modifications can modulate the expression of specific genes including stress-responsive genes [16][17]. These tRNA modification tunable transcripts (MoTTs) respond to the proportion of modified tRNAs and regulate translation in response to cellular stress [18].
–
Threonyl-carbamoyl-adenosine (t6A) is a complex universal modification found at position 37 of nearly all ANN decoding tRNAs, as shown in Figure 1B (for an in depth review of t6A synthesis in all domains of life, see Thiaville et al. [19]). t6A is formed in a two-step mechanism, where, in the cytoplasm of eukaryotes, the threonyl-carbamoyl-AMP (TC-AMP) intermediate is produced by Tcs1 or Tcs2 (previously named YrdC and Sua5, respectively) [20][21][22]. TC-AMP is placed on tRNA by the threonyl-carbamoyl transferase complex (TCTC, previously referred to as KEOPS or EKC complex) made up of Tcs3 (Kae1), Tcs5 (Bud32), Tcs6 (Pcc1), and Tcs7 (Cgi121), whereas fungi have an additional member Tcs8 (Gon7)] [23][24][25]. Yeast mitochondria use a minimum synthesis system to produce t6A-modified tRNAs, consisting of a mitochondrial-targeted Tcs2 and Tcs4 (Qri7) [26][27].
–
In yeast, the absence of t6A synthesis enzymes has been linked to many phenotypes including telomere shortening [28][29], transcription regulation defects [30], and respiration deficiency [31][32][33]. The molecular basis for these pleiotropic phenotypes is far from understood, although it is expected that they should relate to translational defects in absence of t6A. In addition to the aberrant mis-initiation observed in the TCS2 mutant when the gene was discovered [34], the deletion of the TCS2 and TCS3 results in an increase in +1 and -1 frameshifts, as well as to mis-initiation at CUG codons of specific reporter genes [20][23]. Further studies linked loss of TCS2 with increases in leaky scanning bypass of start codons, +1 frameshifts, read-through of UAG, UAA, and UGA stop codons, and an increase in internal ribosome entry site translation (IRES-dependent initiation of translation) [33]. Polysome profiles of TCS2-depleted strains (PTET::TCS2, this strain requires doxycycline for expression of TCS2) revealed abnormal ribosome assembly, which could not be rescued by over-expressing the ternary complex (TC; eIF2α, -β, -γ, and Met-tRNAiMet), contrary to previously reported cases of ribosome assembly defects caused by inhibition of other tRNA modifications [33]. Similarly, the over-expression of either TC or tRNAiMet (IMT4) did not rescue the slow-growth phenotype in absence of t6A [33]. However, depletion of TCS2 leads to increased levels of the transcriptional activator GCN4, although in a non-canonical manner (Gcd– phenotype) [33]. GCN4 is a positive regulator of genes expressed during amino-acid starvation, and is dependent on eIF2α phosphorylation by Gcn2, which monitors uncharged tRNAs [35][36]. Over-expression of tRNAiMet or deletion of GCN2 did not reduce the high levels of GCN4 in a TCS2-depletion background [33]. Paradoxically, GCN4 induction in the TCS2-depleted strain was independent of Gcn2 phosphorylation [33]. In yeast, Gcn4 is also regulated at the translational level by four upstream open reading frames (uORFs), where the scanning ribosome initiates translation at the first AUG in the uORF leading to bypass of initiation at the AUG of the downstream ORFs [37]. TCS2-depletion led to increased translation of the main ORF (GCN4) by bypassing the regulatory uORFs [33]. Over-expression of TC or tRNAiMet did not reduce the leaky scanning seen in TCS2-depletion [33]. Interestingly, mutations of Tcs3, Tcs5, and Tcs8 in yeast also increased GCN4 translation [38].
–
Evidence has emerged that some tRNA modifications can act as determinants of subsequent tRNA modification enzymes. Recently, the requirements of 2’-O-methylation of C32 and N34 has been linked to efficient wybutosine formation at m1G37 of tRNAPhe, a circuitry conserved from yeast to man [39][40][41]. Additionally, in bacteria, presence of the t6A modification increases the efficiency of formation of the essential modification lysidine at U34 of tRNAIleCAU, and t6A is required for the charging of tRNAIle by IleRS [42][43]. In yeast, parallels can also be made between t6A and 5-methoxycarbonylmethyliouridine (mcm5U) and its thiolated derivative (mcm5s2U) found at position 34 of several tRNAs. Both t6A and mcm5s2U modify tRNALysUUU, and mcm5U and t6A are found on tRNAArgUCU [44]. Trm9 and Elp1-6 (Elongator complex) synthesise the mcm5 moiety, and the Ncs2/Ncs6 enzymes are responsible for thiolation [45][46]. Deficiencies in mcm5s2U synthesis lead to slow growth, the inability to grow on non-fermentable carbon sources, and telomere shortening [45][47][48][49][50][51], which are similar to phenotypes seen in t6A biosynthesis mutants [20][23][29][33][34][38][52][53]. Over-expression of a single tRNA, tRNALysUUU, suppresses all of the mcm5s2U phenotypes, and additional data suggest that mcm5s2U acts as a codon-dependent regulator of translation [52][53]. Why elimination of mcm5s2U or t6A lead to similar phenotypes is unknown [2]. One possibility is that the modification of A37 to t6A is required for the formation of the x5s2U derivatives, or vice versa, which has never been explored to date.
–
Recently, both mcm5s2U and t6A have been associated to alterations of two central cell regulatory systems; the General Amino Acid Control system (GAAC) through activation of GCN4 [38][54], which regulates > 1500 genes in response to nutritional cues [35], and Target of Rapamycin Complex (TORC), through alterations in Tor kinase activity [55][56][57][58] (reviewed in Thiaville and de Crécy-Lagard [59]). Modulating the levels of t6A in Drosophila through expression of an unmodifiable tRNAiMet or overexpression of TCS3 led to alterations of Tor activity and changes in whole organism growth [56]. Additionally, knock-down of Tcs3 (Kae1) or Tcs5 (Bud32) in Drosophila larvae activated the Unfolded Protein Response (UPR) [55].
–
Recent ribosome profiling studies of mutations in the mcm5s2U pathway (ncs6∆ and uba4∆) grown under nutrient-depleted conditions revealed pausing and accumulation of ribosomes at GAA, AAA, and CAA codons [54]. Follow up studies also found codon-specific ribosome pausing in the absence of mcm5s2U (ncs2∆elp6∆), even in the absence of stress [60]. Hypo-modified tRNAs cause slower decoding at GAA, AAA, and CAA codons that led to protein misfolding and aggregation of essential proteins, which prevent the cell from maintaining protein homeostasis during stressful events [60].
–
In this study, we sought to uncover the translational defects seen in a t6A deficient strain and to determine if there is a relationship between mcm5s2U and t6A. So far, the links between t6A and translation fidelity have been based on single gene reporters. Here, we sought to assess the functional impact of t6A at the genome scale, by combining genome-wide ribosome profiling and bioinformatics tools to catalogue all translational ambiguities in tcs2∆ and investigate the differential effects of t6A on distinct tRNAs.
RESULTS
mcm5s2U34 or t6A37 are not determinants for each other’s synthesis
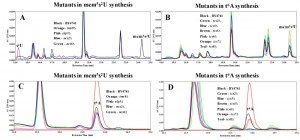
The similarity of the phenotypes observed in strains deficient in mcm5s2U and t6A synthesis suggests that one of the modifications could be required for the synthesis of the other. To test this hypothesis, tRNAs from wild type (BY4741), mcm5s2U-deficient yeast strains (elp3∆, trm9∆, ncs2∆, and ncs6∆) and t6A synthesis mutants (tcs2∆-tcs8∆) were purified and analysed by HPLC.
–
To determine how t6A synthesis deficiency affects mcm5s2U, HPLC analysis with detection at 313 nm (for detection of thio moieties) of nucleosides of tRNAs purified from elp3∆, trm9∆, ncs2∆, and ncs6∆ revealed the mcm5s2U peak at 24.35 minutes, which was unique to BY4741 but absent in all the mutants, and a peak at 14.20 minutes appeared only in elp3∆, indicating the presence of the s2U moiety in this strain (Figure 2A). The chromatographic patterns match previously published reports [50]. Analysis of tRNAs purified from t6A biosynthesis mutants (tcs2∆-tcs8∆) revealed that all strains possessed the peak at 24.35 minutes corresponding to mcm5s2U, and none of the mutants showed the s2U peak at 14.20 minutes (Figure 2B). Interestingly, most mutants in t6A synthesis had higher levels of mcm5s2U (tcs6∆ is unchanged) as compared to BY4741, among which tcs7∆ was the highest (Figure 2B).
The HPLC profile at 254 nm revealed that all the mutants in mcm5s2U synthesis contained the same amount of t6A as the parental BY4741 strain, as indicated on Figure 2C by a peak at 23.57 minutes. Analysis of the t6A synthesis mutants (tcs2∆-tcs8∆) confirmed prior results of the absence of t6A in tcs2∆ and tcs3∆ [20][23], and revealed the absence of t6A in both tcs5∆ and tcs8∆ (Figure 2D). tcs6∆ and tcs7∆ were reduced for t6A relative to wild type by ~20% (Figure 2D), very similar to the reduction seen in a tcs6∆ mutant in the archaea Haloferax volcanii [61]. These results indicate that mcm5s2U34 and t6A37 do not require one another for their synthesis, although eliminating t6A did increase levels of mcm5s2U.
–
Overexpression of tRNAs or Ternary Complex (TC) do not suppress the growth defects of tcs2∆
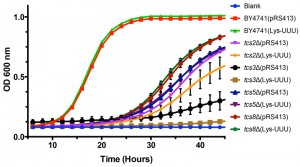
Overexpression of tRNALysUUU is sufficient to suppress all the phenotypes resulting from mutations of mcm5s2U synthesis enzymes [53]. Therefore, we tested if this was also the case for mutations in the t6A synthesis pathway. To assess if tRNAs could suppress the slow growth rate seen in mutants of t6A synthesis, an expression plasmid containing tRNALysUUU was transformed into BY4741, tcs2∆, tcs3∆, tcs5∆, and tcs8∆ (tcs6∆ and tcs7∆ were not tested as they do not have a growth defect). Unlike in the case of mcm5s2U, the expression of tRNALysUUU did not suppress the growth defect observed in t6A synthesis mutants (Figure 3). Interestingly, it led instead to a small reduction in growth rate in the mutants (Figure 3). Further, we expressed the other tRNAs that decode ANN codons (tRNALysCUU, tRNAeMetCAU, tRNAIleAAU, tRNAIleUAU, tRNAThrUGU, tRNAArgACG, tRNAArgUCU, tRNAArgCCU, and tRNAGluUUC (does not contain t6A and decodes GAA, which is the most frequently used codon in S. cerevisiae)) in tcs2∆. None, of these individual tRNAs suppressed the growth defect of tcs2∆ (data not shown). To confirm the results of TCS2-depletion published by Lin et al. [33], plasmids over-expressing tRNAiMet, eIF2α, or TC were transformed into BY4741 and tcs2∆. In agreement with the previous results [33], neither tRNAiMet, eIF2α, nor TC suppressed the slow growth of tcs2∆ (Figure S2A), while the growth of tcs2∆ can be restored by expressing TCS2 in trans (Figure S2B).
Hence, unlike the suppression of mcm5s2U by tRNALysUUU, neither the overexpression of each ANN-tRNA, nor the overexpression of TC components could suppress the fitness defects observed in mutants of the t6A biosynthesis pathway. The effects of the loss of t6A thus appear to be more complex than those of the loss of mcm5s2U.
–
t6A-deficient strains are sensitive to heat and inhibitors of TOR, but growth can be partially rescued by L-homoserine
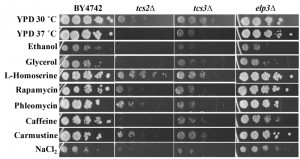
To better characterize how the absence of t6A was affecting cellular function, growth on several carbon sources and under different stress conditions was tested (Figure 4). t6A-deficient strains were found to be sensitive to heat stress, with tcs2∆ unable to grow at 37°C, and to salt stress, with both tcs2∆ and tcs3∆ affected by the presence of 1 M NaCl2. Also, tcs2∆ was unable to grow on 3% glycerol or 6% ethanol, but did grow slowly on 2% glucose (YPD), while tcs3∆ was able to grow slowly on all carbon sources (Figure 4). Addition of inhibitors of the TOR pathway such as caffeine (10 mM) [62] or rapamycin (10 nM) further reduced the growth of t6A-deficient strains (Figure 4). Interestingly, the addition of L-homoserine (1 mg/ml) partially suppressed the growth defects of the tcs2∆ strain, but not of the tcs3∆ strain. Several other chemical stresses did not affect growth of t6A– strains (Figure 4). These included the addition of DNA damaging agents such as phleomycin (8 μg/ml) or carmustine (1 mM). The elp3∆ strain was also tested in the same conditions as it is known that depending on the strain background, mutants in Elongator genes vary as to the degree of response to each of these stressors [63]. The results presented in Figure 4 are consistent with the results recently published by the Schaffrath laboratory, reporting the lack of strong phenotypes of the elp3∆ strain when using the BY4741 background [64].
Homoserine acts as a toxic threonine analogue and incorporation of homoserine activates protein degradation pathways [65]. To test whether activation of UPR could suppress the growth phenotype of t6A-depletion strain in yeast, t6A- strains were transformed with plasmids expressing UPR factors Xbp1, Kar2 (GRP78/BIP), Der1, and Hrd1. Expression of UPR-related factors were unable to rescue the slow-growth phenotype seen in t6A-deficient strains (data not shown).
–
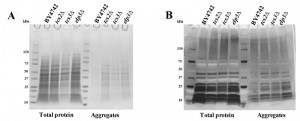
t6A– strains accumulate aggregated proteins and advanced glycated end-products (AGEs)
Double mutants elp6∆ncs2∆ (eliminating mcm5s2U) have been shown to contain increased amount of aggregated (insoluble) proteins, possibly due to alteration in translation speed [60]. Equal amounts of total and insoluble proteins from BY4742, tcs2∆, tcs3∆ and elp3∆ were analysed by SDS-PAGE and Coomassie staining. Depletion of t6A in tcs2∆ and tcs3∆ increased the amount of aggregated or insoluble proteins similar to the single elp3∆ strain (Figure 5A), which is less than the amount of insoluble protein seen in the double elp6∆ncs2∆ strain [60]. Prior experiments in E. coli and H. volcanii revealed that AGEs become more abundant when t6A levels are reduced [61][66]. To assess levels of AGEs in our context, equal amounts of total and insoluble proteins from BY4742, tcs2∆, tcs3∆ and elp3∆ were separated by SDS-PAGE and visualized with a diol-specific silver stain for glycated proteins [67]. Aggregated proteins extracted from tcs2∆, tcs3∆, and elp3∆ were all increased in AGEs relative to wild type (Figure 5B).
Ribosome assembly defects are observed in the t6A– strain
In light of the diverse phenotypes observed and because the previous analysis of translation defects had focussed on a handful of reporter proteins, we performed a global ribosome profiling analysis to assess the impact of t6A-deficiency at the genome scale. An essential step in ribosome profiling is ensuring high quality polysomes are prepared, which was assessed by sucrose gradient sedimentation and subsequent analysis with a fraction analyser. Polysomes prepared from tcs2∆ exhibited a “half-mer” phenotype, which is represented by a shoulder after the 80S peak on the chromatograph, blue arrow in Figure S3. Half-mers indicate excess 40S ribosome and incomplete assembly of the 80S particle, which may indicate problems with initiation [68]. The half-mer phenotype of tcs2∆ was not seen in a prior publication examining a TCS2-depletion strain [33]. These results may differ due to strain genotypes or technical differences in the preparation of the polysomes.
–
The absence of t6A leads to increased ribosome occupancy of arginine synthesis genes
A detailed description of purification of the ribosome-protected fragments (RPFs) and sequencing can be found in material and methods. Analysis of RPFs in tcs2∆ revealed 111 genes were decreased in RPFs and 196 genes were increased in RPFs relative to wild type. A complete list of these genes and their functional roles can be found in Table S3 and Table S4. To determine if any functional relationship existed with these genes, we performed gene ontology (GO) enrichment using YeastMine (http://yeastmine.yeastgenome.org) [69]. The pathway for arginine biosynthesis was found to be enriched, P = 0.049, with five genes, ARG5,6, CPA2, ARG7, ARG1, and CPA1 identified. None of the arginine catabolism pathways were significantly increased (Table S4). Increased mRNA expression of arginine has been documented to act as an anti-oxidant to oxidative stress by an unknown mechanism [70]. This antioxidative pathway acts through pyrroline-5-carboxylate (P5C) but PUT1, encoding a P5C synthesis enzyme, was not increased in RPFs in the mutant (Table S4). Of the 111 genes decreased in RPFs, five genes were identified matching the GO term polyphosphate metabolic process, P = 0.003, and no pathway enrichment was identified.
–
Depletion of t6A deregulates GCN4
The number of genes proposed to be regulated by Gcn4 varies greatly with the specific study, from less than 500 genes (microarrays measuring gene expression during histidine starvation) [35] to more than 2500 genes (predicted computationally by SGD). The most conservative estimate of Gcn4-induced genes was produced from a ChIP-Chip assay, which found 128 genes bound during immunoprecipitation of Gcn4 [71]. Comparison of the RPFs detected in tcs2∆ with the 128 well defined Gcn4-regulated genes reveals that 15 ORFs increased in RPFs are regulated by Gcn4 in tcs2∆, while no ORFs decreased in RPFs in tcs2∆ are regulated by Gcn4, Figure S4A. The 15 Gcn4-regulated genes with increased RPFs in tcs2∆ are involved in amino acid synthesis, with six in the arginine synthesis pathway (P = 1.3 x 10-5), Table S5.
–
Microarray analyses of conditional or point mutations in tcs3, tcs6, or tcs8 had been previously reported [38] and in all these studies, an up-regulation of Gcn4 regulated genes was observed, including genes in the arginine and histidine biosynthesis pathways, although the mRNA expression levels of GCN4 itself did not increase. The genes increased in each of the previous microarray studies were compared to the genes with increased RPFs in the ribosome profiling analysis of tcs2∆. Of the 196 genes with increased RPFs in tcs2∆, 29 were also increased in tcs3-18, 30 were also increased in tcs6-4, and 12 were also increased in tcs18-ts1, summarized in Figure S4B. 12 genes were increased in all four datasets (Table 1). 9 of these are under Gcn4 control, of which four are in the arginine synthesis pathway (Table 1).
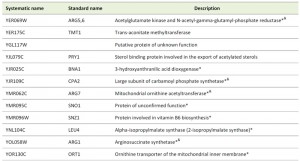 | Table 1. Genes increased in expression in tcs2∆, tcs3-18, tcs6-4, and tcs8-ts1. *Under control of Gcn4 (see Table S5), &Arginine biosynthesis. |
Contrary to the previous microarray results that did not detect GCN4 induction, we found that RPFs mapping to GCN4 were increased 6-fold in tcs2∆ (Table S4). This difference may be due to an up-regulation of translation (detected by the increased levels of RPFs in tcs2∆), and not transcription (as measured by the microarrays). With the 6-fold increase in GCN4 expression, it is surprising so few Gcn4 inducible genes are increased in tcs2∆. Indeed, only 8% of RPFs increased in tcs2∆ are in common with the Gcn4p ChIP data.
–
A discrete but not a global increase in translational ambiguity is observed in the t6A– strain tcs2∆
In yeast, the RPF is 28 nucleotides long [72], hence to analyse the frame of each ribosome, only 28-mers that aligned uniquely to the genome and did not contain mismatches were used. 6.5 x 106 reads in wild type and 5.8 x 106 reads in tcs2∆ matched these strict criteria. For each read, the nucleotide at position +12, which corresponds to the ribosomal P-site, was determined and its identity defined the frame of the read, and hence the frame of the ribosome, Figure S5. Each ORF was divided into windows of approximately 300 nucleotides (minimum of 3 windows per ORF to a maximum of 9), and reads inside each window were mapped and enumerated [73]. Since we cannot be sure if the ribosome associated with read is in frameshift or if that ribosome began translation of the ORFs out of frame, translational ambiguity is defined as the mapping of a read in a frame other than the frame of the annotated ORF.
–
There are four well-documented examples of +1 frameshifts occurring in S. cerevisiae and these were used to evaluate the frame analysis performed here. One known +1 frameshift, TRM140 (an AdoMet-dependent tRNA methyltransferase), was detected in both BY4742 and tcs2∆ and is illustrated in Figure S6A and B. For translation of full-length Trm140, the ribosome must undergo a +1 frameshift at nucleotide 832. As seen in Figure S6A and B, nearly 100% of the reads begin in Frame 0, then after base 832, nearly 100% of the reads are in the +1 frame.
–
87 and 213 ORFs were found to have potential translational ambiguities in BY4742 and tcs2∆, respectively (Table S6 and Table S7). GO term enrichment of these genes with translational ambiguities revealed a single biological process, cytoplasmic translation, was enriched in both strains, with 16 genes in BY4742 (P = 4 x 10-6) and 35 genes in tsc2∆ (P = 3 x10-11). In tcs2∆, 17 of the 79 ribosomal proteins had increased levels of translation ambiguities (Table S7).
–
Interestingly, global analysis of translational ambiguities, by summing all reads used to determine frame, indicates that 80% of all reads from the ribosome profiling are in the correct, annotated frame (Frame 0), but there is a significant difference in translational ambiguities between wild type and mutant (P = 6.5 X 10-98, t-test), Figure S7. Interestingly, only 0.26% of all ORFs were identified as having potential translational ambiguities in tcs2∆. Thus, the data indicates that loss of t6A is causing ambiguities at discrete sequences, or codons, but is not causing a global, cataclysmic alteration of reading frame.
–
The number observed non-AUG starts doubles in the t6A-deficient strain
TCS2 (SUA5) was discovered in yeast as a suppressor of a translational initiation defect in the cyc1-362 allele [34]. cyc1-362 contains an aberrant upstream and out of frame AUG resulting in ~2% of the normal Cyc1 protein levels. Suppressors would bypass the out of frame AUG and initiate at the correct downstream AUG, increasing the amount of Cyc1 [34]. To detect initiation of translation at non-conical codons, we parsed the profiling data with a strict set of parameters. To be considered a non-canonical start codon, a GUG, UUG, or GUC codons (the most frequently used non-AUG initiation codons in yeast) [74][75][76][77][78] had to be within 100 nucleotides upstream of the ORF of interest, and be in-frame with the downstream AUG with no stop codon between the candidate non-AUG and its downstream AUG. Finally, a minimum of 128 reads was required to cover the non-AUG site.
–
In yeast, there are two well-characterized occurrences of non-AUG initiation occurring upstream of the annotated AUG start site. ALA1 encodes both the cytoplasmic and mitochondrial alanyl-tRNA synthetase. The cytoplasmic form of Ala1p is translated from the annotated AUG, and the mitochondrial form is translated from a pair of ACG codons located at -25 and -24 relative to AUG [79]. GRS1 encodes the cytoplasmic and mitochondrial glycyl-tRNA synthase. The cytoplasmic form of Grs1p is translated from the annotated AUG, and the mitochondrial form is translated from a UUG codon located at -26, relative to the AUG [78]. In both BY4742 and tcs2∆, initiation at the upstream non-AUG codons can be detected for ALA1 and GRS1, Figure S8A and Figure S8B.
–
The analysis of non-AUG initiation was expanded to the entire profiling dataset. For the three codons analysed, tcs2∆ contain nearly twice as many non-AUG starts as BY4742. For initiation at UUG, BY4742 contained 140 genes, Table S8, and tcs2∆ contained 260, Table S9. For initiation at ACG, BY4742 contained 98 genes, Table S10, and tcs2∆ contained 169, Table S11. For initiation at GUG, BY4742 contained 62 genes, Table S12, and tcs2∆ contained 134, Table S13. None of these sets of genes contained any enrichment of GO terms.
–
t6A’s role in translation speed varies with the codon
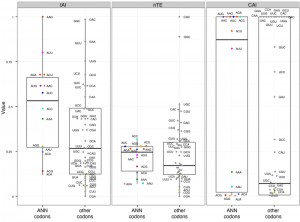
Different metrics have been developed to estimate translation efficiencies of individual codons based on the abundance of their cognate tRNAs, and the properties of the ASL they form. There are three major metrics commonly used to measure the translation efficiency of codons: the Codon adaptation index (CAI, [80]), the tRNA adaptation index (tAI, [81]), and the normalized Translational Efficiency (nTE, [82]) (which is based on codon abundance in the transcriptome rather than codon frequency in the genome). Using the three metrics, we found that ANN codons in yeast have in average a higher estimated translational efficiency compared to other codons (Figure 6), suggesting that there is a statistical tendency for ANN-tRNAs to be in high supply in standard growth conditions. The predicted efficiencies vary greatly between ANN codons, with AAG always in high supply while AUA always in low supply (Figure 6).
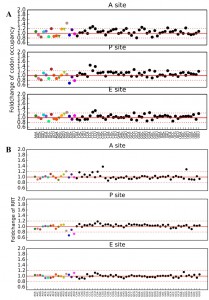
Ribosome profiling data allows for evaluation of translation speed by measuring codon occupancy at each site in the ribosome, with increased occupancy analogous to a decrease elongation rate, and vice versa. Using two different methods, the Codon Occupancy (CO) [83] and the Ribosome residence time (RRT) [84], a count of every codon occupying the ribosomal A (acceptor), P (peptidyl transfer), and E (exit) sites was compiled. Comparing the Log2 fold-change of CO and RRT in the A, P, and E sites of tcs2∆ and BY4742 produced a global summary of the consequences of t6A absence on decoding, Figure 7.
The ribosomal A site occupancy for t6A dependent codon site occupancy was increased for AUA, ACG, AGG, AUG, ACA, AAA, and AAU and decreased for AUU, AAC, AUC, AGA, ACU, ACC, and AAG. The codons with increased occupancy in the A-site fell into two categories: (i) codons that are decoded by rare tRNAs (only 1-4 copies of tRNAs genes are encoded in the chromosome) and (ii) codons that are decoded by a G34:U3 wobble, as for AAU (decoded by tRNAAsn(GUU)) (see Figure 1B for codon:anticodon pairs). The codons whose A-site occupancy decreased also fell into two categories: (i) codons decoded abundant by tRNAs (4-13 genes) and (ii) codons decoded by an I34:C3 wobble, as for AUC (decoded by tRNAIle(IAU)) and ACC (decoded by tRNAThr(IGU)) (Figure 1B). The pattern found for A-site occupancy, also held true for P-site and E-site occupancies. Interestingly, this pattern also held true for codons decoded by non t6A-containing tRNAs. AGU (G:U wobble) and CGG (decoded by the rare tRNAArgCCG) were increased in ribosome occupancy, while GUC and GCU (I:C or I:U) were decreased in ribosome occupancy (Figure 7). From this data, it appears that t6A is helping increase elongation rate of rare tRNAs and G34:U3 pairs and decrease the elongation rate of high abundance and I34:C3 pairs to homogenize the process of elongation.
DISCUSSION
The absence of t6A in yeast does not lead to catastrophic and global defects in translation, as would be expected from previous studies based on single reporter assays. Even with doubling of initiation at upstream non-AUG starts and a 2.5 fold increase in translational ambiguities, only a limited number of genes in the yeast genome were affected. This suggests that the severe and pleiotropic phenotypes caused by t6A deficiency may not be caused by global defects in translation, but instead of the subtler consequences of codon-specific translation defects caused by lack of t6A.
–
Role of t6A in decoding efficiency varies with the tRNA
The codon occupancy results presented in this study suggest that t6A helps rare cognate tRNAs and G:U mismatches (near-cognates) compete with Watson:Crick decoding tRNAs and slows decoding by high abundance tRNAs and tRNAs using the wobble U:C base pairings [85]. This can be all the more critical for codons like AGG decoded by tRNAArgCCU that are known to be strongly inhibitory for translation efficiency [86][87][88]. Another important role for t6A is stabilizing the interaction between the first base of the mRNA codon and position 36 (the third nucleoside) of the tRNA anticodon, preventing decoding of near-cognates by tRNAiMetCAU [89]. This is exemplified by tRNAfMet of E. coli that contains an unmodified A37 and can efficiently decode GUG and UUG while the eukaryotic tRNAiMet contains t6A37 and rarely decodes non-AUG codons [90][91][92][93]. The examination of alternative start-sites presented here supports the role of t6A preventing tRNAiMet from recognizing near-cognates and restricting translation initiation to AUG codons.
–
The codon occupancy for non-t6A containing tRNAs is also altered (Figure 7), and this is more dramatic than what is seen in ncs2∆elp6∆ [60]. This could be due to an alteration in competition between cognate and near-cognate tRNAs. This was previously demonstrated for tRNAArgUCU (tRNAArgIII), which naturally exists in t6A modified and unmodified forms [94]. The modified version of tRNAArgUCU can outcompete the unmodified form for the cognate codon and binds more tightly to tRNASerGGA involving a U36:G34 mismatch [94]. CGG codons (decoded by tRNAArgCCG) and UGG codons (tRNATrpCCA) are increased in both the codon occupancy and RRT assays (Figure 7). One can speculate that the slower decoding at CGG and UGG is due to competition between tRNAArgCCG or tRNATrpCCA with an unmodified near-cognate tRNAArgCCU.
–
A recent global analysis of yeast ribosome profiling data has shown that frequent codons are decoded more quickly than rare codons, and AT-rich codons are decoded more quickly than GC-rich codons [84]. It seems that the difference could be even larger if tRNA modifications are altered, as shown here with the absence of t6A, and as is already know for several other ASL modifications. The absence of Queuosine (Q34) is known to have opposite effects on decoding depending on identity of the 3rd base of the codon [88][95], and the depletion of mcm5s2U synthesis can alter the decoding rates of tRNAs that do not possess this modification [60]. An emerging general trend for the ASL modifications is to homogenize the kinetics of individual tRNA binding (competition) during translation and to alter the speed of translation to ensure proper protein folding, a concept that was predicted by Toshimichi Ikemura in 1981 [96].
–
Could defects in translation speed cause the pleiotropic phenotypes of t6A–?
Analysis of codon stretches in yeast [20] revealed that the genes with the longest stretches of t6A-dependent codons encode poly-Asn (poly-N) proteins that contain up to 31 consecutive AAU/AAC codons. These include GPR1, required for glucose activation of the cAMP pathway [97], and SWI1, a subunit of the SWI/SNF chromatin remodelling complex required for transcription of many genes involved in sugar catabolism, as well as meiosis cell mating type (see summary in SGD [98]). If the protein expression of these two genes is reduced in the absence of t6A (that is a decrease in elongation speed of these transcripts possibly due to stalling), many of the phenotypes seen in t6A– strains (e.g., no growth on galactose, chromatin remodelling defects and telomere shortening [33][99][100]) can be explained. Unfortunately, the presence of these repeats (90 nts) are longer than the RPFs (28 nts) sequenced, so these genes could not be analysed in the ribosome profiling and further studies are needed to test this hypothesis.
–
Several stress-induced transcription factors are also increased in tcs2∆ context, including NSF1 (YPL230W), a transcriptional regulator of genes involved in growth on non-fermentable carbon sources (see summary in SGD [98]), Sol4 (YGR248W), which functions in the pentose phosphate pathway (see summary in SGD [98]), and Smc6 (YLR383W), a component of the SMC5-SMC6 complex that plays a key role in the removal of X-shaped DNA structures (see summary in SGD [98]) (Table S4). Up-regulation of any of these transcription factors would have wide-ranging effects and could explain some of the pleiotropic phenotypes seen under t6A deficiency.
–
Ribosome profiling of ncs2∆ revealed genes with increased translation activity tend to play a role in amino acid metabolism, and GCN4 is significantly increased [54]. Comparison of genes increased in tcs2∆ and ncs6∆ revealed 19 genes found in each of these mutants, Table S14. Increased genes included GCN4 and several members of the arginine biosynthesis pathway, Table S14. Twelve genes are decreased in both tcs2∆ and ncs6∆, including two ribosomal protein subunits and two phosphatases, Table S15. GCN4 is also increased in the ribosome profiling of ncs2∆elp6∆ [60]. A common theme seen in disrupting ASL modifications is the de-repression of Gcn4, in a non-canonical Gcn2-independent manner, activating a small subset of Gcn4 regulated genes. The mechanism of activation and the importance of Gcn4 activation are not understood at this point in time.
–
Restoring protein homeostasis suppresses the slow growth of tcs2∆
An explanation for the similarity of phenotypes seen in both Elongator and t6A mutants could be due to the disruption of protein homeostasis. Unlike the mutations in the Elongator modification, the slow growth observed during disruption of t6A biosynthesis cannot be suppressed by overexpression of tRNAs (Figure 3). However, L-homoserine rescued the growth of tcs2∆, but not tcs3∆ (Figure 4). Further studies are required to explain this suppression, but homoserine is a toxic intermediate, which acts as a threonine analogue, and it was recently shown that the ubiquitin pathway and the proteasome are crucial in alleviating homoserine toxicity [65].
–
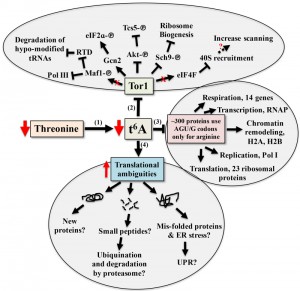
Model for the cellular response to reduction of t6A
To date, tcs2 and members of the TCTC complex have been implicated in transcriptional regulation. While there is no empirical evidence eliminating this possibly, the evidence presented here and in other works suggests transcriptional changes seen when perturbing t6A biosynthetic genes are part of an adaptive response by the cell to cope with translational errors. The response to alterations in t6A levels involves a combination of both independent and interrelated events, summarized in Figure 8. The model proposes that t6A acts as a sensor of nutritional levels as t6A varies in response to the availability of threonine, Figure 8-1 [101]. As t6A levels decline, Tor1 activity decreases, Figure 8-2 [56]. As a master controller, declines in Tor1 activity reduces the growth potential of the cell [55][56][102] and has wide ranging affects, from blocking Pol I, Pol III [103], and the RTD (Rapid tRNA Degradation pathway to prevent degradation of hypo-modified tRNAs) [104] to lessening translation initiation and ribosome biogenesis [105].
A reduction in t6A may also lower the translation rate of specific proteins due to codon usage, Figure 8-3. Arginine is one of two amino acids in yeast that are incorporated both by t6A-containing tRNAs (AGA/G codons) and by tRNAs lacking t6A (CGN codons), Figure 1B [44]. The AGA/G codons are known to be frequent sites of frameshifting, reviewed in [106]. Using the codon usage database [107], yeast genes were ranked according to their use of t6A-dependent or t6A-independent Arg codons. Around 300 genes used only t6A-dependent Arg codons and GO analysis showed a strong enrichment for genes of the aerobic respiration and electron transport pathways (P – value 10-9, Holm-Bonferroni test), which could explain the respiratory deficiency phenotype displayed by t6A– yeast strains (Table S16 and S17). 23 ribosomal proteins, RNA Polymerase subunits (RNAP), and proteins of the chromatin remodelling complexes H2A and H2B use t6A-dependent Arg codons. Only 12 of these t6A-dependent Arg genes were decreased in RPFs in tcs2∆. Proteomic studies are now underway to confirm if this specific set of genes are translated less efficiently in the t6A– strains.
–
As t6A levels decrease, translation fidelity decreases (Figures S7, S8 and Tables S6-13). Increase in translation ambiguity could lead to new protein products, which may be non-functional or toxic [108]. Out-of-frame decoding could increase synthesis of small peptides [108] and misfolded proteins could lead to the activation of the unfolded protein response (UPR) [55], and to the activation of catabolic pathways [109][110], Figure 8-4. Additional proteomic studies are underway to measure misfolding and amino acid misincorporation rates in t6A– strains to further characterize these multi-layered and complex phenotypes.
MATERIALS AND METHODS
Strains and growth conditions
A list of all organisms used in this study can be found in Table S1. Yeast strains were grown on YPD (DIFCO Laboratories) at 30°C. Synthetic minimal media (SD), with or without agar, with or without dropout supplements (-uracil, -ura; -leucine, -leu; -histidine, -his) were purchased from Clontech (Palo Alto, CA) and prepared as recommended by the manufacturer. Glucose (Glu, 2% w/v), Glycerol (Gly, 4% w/v), Ethanol (EtOH, 6% v/v) 5-fluoro-orotic acid (5-FOA, 0.1% w/v) and G418 (300 mg/mL) were used when appropriate. Yeast transformations were carried out using frozen competent cells as described by [111] with plating onto the appropriate media. The S. cerevisiae tcs2∆::KanMX4 strain, VDC9100, was created as previously described [27]. All strains were genotyped using oligonucleotides targeting inside and outside the gene of interest, in addition to the location of the replacement cassette. Oligonucleotides are listed in Table S2. VDC9100 (tcs2∆) harboring tRNA over-expression plasmids were created using the plasmid shuffle technique by first transforming with pBN204 (TCS2 complementation plasmid), then transforming with tRNA plasmids, and finally curing pBN204 from VDC9100 using SD-leu+5-FOA media.
–
Yeast growth assays
Growth curves were performed using a Bioscreen C MBR (Oy Growth Curves AB Ltd, Finland) at 30°C and at maximum shaking. A 250 µl culture was used in each well, and 5 biological replicates were used for each condition. Yeast cultures were grown in the listed media to saturation, normalized to an OD600 of 1, and diluted 200 times in the listed media before loading on the Bioscreen. The growth curves presented are averages of 5 biological replicates. Significance was determined using a 2-way ANOVA and Fisher’s LSD using Prism 6 (GraphPad).
–
For phenotype screens and tRNA over-expression assays, yeast cultures were grown in the media listed in the figure to saturation, normalized to an OD600 of 1.0 and 5 µL of 1:10 serial dilutions were spotted on the listed media with the supplements listed in the figure and text. Galactose (2% w/v) was added when needed.
–
Extraction and digestion of bulk tRNAs
Bulk tRNAs were prepared as previously described using acid buffered-phenol (phenol saturated with 50 mM sodium acetate, pH 5.8) and alcohol precipitation [20]. Nucleosides were prepared as described in [61] by hydrolyzing bulk tRNA with 10 units of Nuclease P1 (Sigma) overnight at 37°C, with the addition of 0.01 units of phosphodiesterase I (Sigma) and 3 µL E. coli alkaline phosphatase (Sigma). The hydrolyzed nucleosides were further purified by filtering through a 5 kD MWCO filter (Millipore) (to remove enzymes), dried in a CentriVap Concentrator, and suspended in 20 µL of water prior to analysis by HPLC or LC-MS/MS.
–
HPLC and LC-MS/MS Analysis
t6A was detected by HPLC as described by [112] using a Waters 1525 HPLC with Empower 2 software and detected with a Waters 2487 UV-vis spectrophotometer with simultaneous detection at 254 nm and 313 nm (for thio-derivatives). Separation was performed on an Ace C-18 column heated to 30°C, using 250 mM ammonium acetate (Buffer A) and 40% acetonitrile (Buffer B) run at 1 mL/min. 100 µg of nucleosides were injected and separated using a complex step gradient [112]. Levels of t6A were measured by integrating the peak area from the extraction ion chromatograms. The ratio of Ψ-modified base/m22G was used to normalize for tRNA concentration across samples. Levels for mutant strains were expressed relative to wild type levels. Results were confirmed by LC–MS/MS at the Donald Danforth Plant Science Center, St. Louis MO, as described in [20]. The MS/MS fragmentation data, as well as a t6A standard provided by D. Davis (University of Utah) were also used to confirm the presence of t6A.
–
Ribosome profiling
Purification of RPFs and library preparation
Ribosome Profiling was performed as described previously by Baudin-Baillieu et al. [113][114]. Briefly, polysomes were prepared from two biological replicates of the parental BY4742 strain and tcs2∆ (VDC9100) grown from a preculture diluted into 500 mL YPD in an Erlenmeyer flask. Cells were harvested at OD600 0.6, chilled on ice, and cycloheximide was added to a final concentration of 50 µg/mL. Polysomes were harvested in cold lysis buffer (0.1 mM Tris-HCl, pH 7.4, 10 mM NaCl2, 3 mM MgCl2, and 50 µg/mL cycloheximide), and were aliquoted at approximately 40-50 OD260 units per tube and rapidly frozen in liquid nitrogen and stored at -80°C. Monosomes were prepared by digesting polysome extracts for 1 hour at room temperature with 15 units RNaseI (Ambion) per OD unit. The digested polysomes were purified on sucrose gradients prepared by casting the sucrose gradients (31% sucrose, 50 mM Tris-acetate pH 7.6, 50 mM NH4Cl, 12 mM MgCl2, and 1 mM DTT) with three freeze-thaw cycles. The samples were loaded on the gradients and centrifuged in a Beckman SW41 rotor at 39,000 rpm at 4°C for 3 hours. Fractions were collected using an ISCO (Teledyne, Lincoln, NE) instrument at a 0.5 mL/min flow rate. Ribosome protected fragments (RPFs) of mRNA were purified using acid phenol (unbuffered), chloroform, and ethanol precipitation, then stored at -20°C. The 28-nucleotide RNA fragments were selected on 15% acrylamide gels containing 7 M urea. A 28 nt RNA oligonucleotide (oNTI 199 5′-AUGUACACGGAGUCGACCCGCAACGCGA-3′) was used as a size marker. After migration, gels were incubated for 30 minutes in a 10 % solution of SYBR-Gold (Life Technologies) and visualized with a UV lamp at 300 nm, and the 28 nt fragments were excised. The excised gel pieces were loaded into the pierced 1.5 mL tubes inside a 2 mL tube, and centrifuged for 1 minute at 16,000 x g. RNA was precipitated with glycogen ethanol overnight at -20°C. RPFs were depleted of major rRNA contamination by subtractive hybridization using biotinylated oligonucleotides (Table S2) and were recovered by reacting with MageneShere Paramagnetic streptavidin particles (Promega). The supernatants containing the RPFs were recovered and the RNA was precipitated, as described above. RNA size and quality was checked with a Small RNA Chip on a Bioanalyzer 2100 (Agilent). A directional RNA-Seq library was prepared by IMAGIF (Centre de Recherche de Gif – http://www.imagif.cnrs.fr) Gif-sur-Yvette, France using the TruSeq Small RNA Sample Prep Kit (Illumina) and the v1.5 sRNA 3′ Adaptor (Illumina) according to the manufacturer’s protocol and verified using Bioanalyzer Small RNA Analysis kit (Agilent). Sequencing was performed at the Microarray and Genomic Analysis Core Facility at the University of Utah Huntsman Cancer Institute on an Illumina HiSeq 1500 and subjected to a 50 cycle run.
–
Sequencing, quality control, and read-mapping
Sequencing and bioinformatics analysis were performed as described in [114]. In brief, four sequencing libraries were prepared from the 28-mer RPFs purified from two biological replicates of BY4742 and tcs2∆. The libraries were sequenced on an Illumina HiSeq 1500 with 50-cycle single-end reads to maximize the numbers of reads for each small RNA library. Approximately 2 x 109 reads were obtained for each sample. Quality was assessed using FastQC http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and adaptors were removed using CutAdapt [115]. To remove contaminating reads corresponding to rRNA, reads were mapped against an rRNA index from the Saccharomyces Genome Database (SGD) using Bowtie2 with the default settings [116]. Reads not mapping to rRNA were mapped against the SacCer3 index (SGD) [117] using Bowtie2. Approximately 1.3 x 108 reads from each sample were mapped to the S. cerevisiae genome. Differential expression between samples was determined using a DESeq, with multiple testing correction using Benjamini and Hochberg [118][119][120]. Significant differences between wild type and mutant were declared based on an adjusted alpha of 0.05. Precision of the biological replicates was very high with R = 0.9881 and R = 0.9959 for BY4742 and tcs2∆, respectively (Figure S1A and B). Post-sequencing analysis to identify differential expression, frameshifts, read-through, and non-AUG starts was performed as described in [73]. Sequences were deposited at NCBI GEO database under accession number GSE72030.
–
Functional classification of genes
Lists of genes produced from the above analysis were analysed using YeastMine [69], an interactive database for querying the Saccharomyces Genome Database (SGD, http://www.yeastgenome.org) [69] to produce Gene Ontology enrichments and pathway enrichments.
–
Detection of protein aggregates and AGEs
Proteins were extracted from cells grown to mid-log and total proteins were extracted as described in [121]. Aggregates were isolated as described by [122]. Total proteins and aggregates were separated on 4-20% denaturing polyacrylamide gels with Coomasie blue staining. AGEs were identified by diol-specific silver staining [67].
References
- F. Crick, "Codon—anticodon pairing: The wobble hypothesis", Journal of Molecular Biology, vol. 19, pp. 548-555, 1966. http://dx.doi.org/10.1016/S0022-2836(66)80022-0
- B. El Yacoubi, M. Bailly, and V. de Crécy-Lagard, "Biosynthesis and Function of Posttranscriptional Modifications of Transfer RNAs", Annual Review of Genetics, vol. 46, pp. 69-95, 2012. http://dx.doi.org/10.1146/annurev-genet-110711-155641
- H. Grosjean, V. de Crécy-Lagard, and C. Marck, "Deciphering synonymous codons in the three domains of life: Co‐evolution with specific tRNA modification enzymes", FEBS Letters, vol. 584, pp. 252-264, 2009. http://dx.doi.org/10.1016/j.febslet.2009.11.052
- E. Novoa, M. Pavon-Eternod, T. Pan, and L. Ribas de Pouplana, "A Role for tRNA Modifications in Genome Structure and Codon Usage", Cell, vol. 149, pp. 202-213, 2012. http://dx.doi.org/10.1016/j.cell.2012.01.050
- T.N. Lamichhane, N.H. Blewett, A.K. Crawford, V.A. Cherkasova, J.R. Iben, T.J. Begley, P.J. Farabaugh, and R.J. Maraia, "Lack of tRNA Modification Isopentenyl-A37 Alters mRNA Decoding and Causes Metabolic Deficiencies in Fission Yeast", Molecular and Cellular Biology, vol. 33, pp. 2918-2929, 2013. http://dx.doi.org/10.1128/MCB.00278-13
- E.M. Novoa, and L. Ribas de Pouplana, "Speeding with control: codon usage, tRNAs, and ribosomes", Trends in Genetics, vol. 28, pp. 574-581, 2012. http://dx.doi.org/10.1016/j.tig.2012.07.006
- W. Ran, and P.G. Higgs, "The Influence of Anticodon-Codon Interactions and Modified Bases on Codon Usage Bias in Bacteria", Molecular Biology and Evolution, vol. 27, pp. 2129-2140, 2010. http://dx.doi.org/10.1093/molbev/msq102
- W. Ran, D.M. Kristensen, and E.V. Koonin, "Coupling Between Protein Level Selection and Codon Usage Optimization in the Evolution of Bacteria and Archaea", mBio, vol. 5, 2014. http://dx.doi.org/10.1128/mBio.00956-14
- W. Ran, and P.G. Higgs, "Contributions of Speed and Accuracy to Translational Selection in Bacteria", PLoS ONE, vol. 7, pp. e51652, 2012. http://dx.doi.org/10.1371/journal.pone.0051652
- J.B. Plotkin, and G. Kudla, "Synonymous but not the same: the causes and consequences of codon bias", Nature Reviews Genetics, vol. 12, pp. 32-42, 2010. http://dx.doi.org/10.1038/nrg2899
- A. Krisko, T. Copic, T. Gabaldón, B. Lehner, and F. Supek, "Inferring gene function from evolutionary change in signatures of translation efficiency", Genome Biology, vol. 15, pp. R44, 2014. http://dx.doi.org/10.1186/gb-2014-15-3-r44
- E.W. Wallace, E.M. Airoldi, and D.A. Drummond, "Estimating Selection on Synonymous Codon Usage from Noisy Experimental Data", Molecular Biology and Evolution, vol. 30, pp. 1438-1453, 2013. http://dx.doi.org/10.1093/molbev/mst051
- D. Allan Drummond, and C.O. Wilke, "The evolutionary consequences of erroneous protein synthesis", Nature Reviews Genetics, vol. 10, pp. 715-724, 2009. http://dx.doi.org/10.1038/nrg2662
- D.A. Drummond, J.D. Bloom, C. Adami, C.O. Wilke, and F.H. Arnold, "Why highly expressed proteins evolve slowly", Proceedings of the National Academy of Sciences, vol. 102, pp. 14338-14343, 2005. http://dx.doi.org/10.1073/pnas.0504070102
- P.C. Dedon, and T.J. Begley, "A System of RNA Modifications and Biased Codon Use Controls Cellular Stress Response at the Level of Translation", Chemical Research in Toxicology, vol. 27, pp. 330-337, 2014. http://dx.doi.org/10.1021/tx400438d
- M.A. Preston, S. D’Silva, Y. Kon, and E.M. Phizicky, "tRNAHis 5-methylcytidine levels increase in response to several growth arrest conditions in Saccharomyces cerevisiae", RNA, vol. 19, pp. 243-256, 2012. http://dx.doi.org/10.1261/rna.035808.112
- C.T.Y. Chan, M. Dyavaiah, M.S. DeMott, K. Taghizadeh, P.C. Dedon, and T.J. Begley, "A Quantitative Systems Approach Reveals Dynamic Control of tRNA Modifications during Cellular Stress", PLoS Genetics, vol. 6, pp. e1001247, 2010. http://dx.doi.org/10.1371/journal.pgen.1001247
- C. Gu, T.J. Begley, and P.C. Dedon, "tRNA modifications regulate translation during cellular stress", FEBS Letters, vol. 588, pp. 4287-4296, 2014. http://dx.doi.org/10.1016/j.febslet.2014.09.038
- P.C. Thiaville, D. Iwata-Reuyl, and V. de Crécy-Lagard, "Diversity of the biosynthesis pathway for threonylcarbamoyladenosine (t6A), a universal modification of tRNA", RNA Biology, vol. 11, pp. 1529-1539, 2014. http://dx.doi.org/10.4161/15476286.2014.992277
- B. El Yacoubi, B. Lyons, Y. Cruz, R. Reddy, B. Nordin, F. Agnelli, J.R. Williamson, P. Schimmel, M.A. Swairjo, and V. de Crecy-Lagard, "The universal YrdC/Sua5 family is required for the formation of threonylcarbamoyladenosine in tRNA", Nucleic Acids Research, vol. 37, pp. 2894-2909, 2009. http://dx.doi.org/10.1093/nar/gkp152
- C. Deutsch, B. El Yacoubi, V. de Crécy-Lagard, and D. Iwata-Reuyl, "Biosynthesis of Threonylcarbamoyl Adenosine (t6A), a Universal tRNA Nucleoside", Journal of Biological Chemistry, vol. 287, pp. 13666-13673, 2012. http://dx.doi.org/10.1074/jbc.M112.344028
- C.T. Lauhon, "Mechanism of N6-Threonylcarbamoyladenonsine (t6A) Biosynthesis: Isolation and Characterization of the Intermediate Threonylcarbamoyl-AMP", Biochemistry, vol. 51, pp. 8950-8963, 2012. http://dx.doi.org/10.1021/bi301233d
- B. El Yacoubi, I. Hatin, C. Deutsch, T. Kahveci, J. Rousset, D. Iwata-Reuyl, A. G Murzin, and V. de Crécy-Lagard, "A role for the universal Kae1/Qri7/YgjD (COG0533) family in tRNA modification", The EMBO Journal, vol. 30, pp. 882-893, 2011. http://dx.doi.org/10.1038/emboj.2010.363
- L. Perrochia, D. Guetta, A. Hecker, P. Forterre, and T. Basta, "Functional assignment of KEOPS/EKC complex subunits in the biosynthesis of the universal t 6 A tRNA modification", Nucleic Acids Research, vol. 41, pp. 9484-9499, 2013. http://dx.doi.org/10.1093/nar/gkt720
- L. Perrochia, E. Crozat, A. Hecker, W. Zhang, J. Bareille, B. Collinet, H. van Tilbeurgh, P. Forterre, and T. Basta, "In vitro biosynthesis of a universal t6A tRNA modification in Archaea and Eukarya", Nucleic Acids Research, vol. 41, pp. 1953-1964, 2012. http://dx.doi.org/10.1093/nar/gks1287
- L.C.K. Wan, D.Y.L. Mao, D. Neculai, J. Strecker, D. Chiovitti, I. Kurinov, G. Poda, N. Thevakumaran, F. Yuan, R.K. Szilard, E. Lissina, C. Nislow, A.A. Caudy, D. Durocher, and F. Sicheri, "Reconstitution and characterization of eukaryotic N6-threonylcarbamoylation of tRNA using a minimal enzyme system", Nucleic Acids Research, vol. 41, pp. 6332-6346, 2013. http://dx.doi.org/10.1093/nar/gkt322
- P.C. Thiaville, B. El Yacoubi, L. Perrochia, A. Hecker, M. Prigent, J.J. Thiaville, P. Forterre, O. Namy, T. Basta, and V. de Crécy-Lagard, "Cross Kingdom Functional Conservation of the Core Universally Conserved Threonylcarbamoyladenosine tRNA Synthesis Enzymes", Eukaryotic Cell, vol. 13, pp. 1222-1231, 2014. http://dx.doi.org/10.1128/EC.00147-14
- F. Meng, Y. Hu, N. Shen, X. Tong, J. Wang, J. Ding, and J. Zhou, "Sua5p a single-stranded telomeric DNA-binding protein facilitates telomere replication", The EMBO Journal, vol. 28, pp. 1466-1478, 2009. http://dx.doi.org/10.1038/emboj.2009.92
- M. Downey, R. Houlsworth, L. Maringele, A. Rollie, M. Brehme, S. Galicia, S. Guillard, M. Partington, M.K. Zubko, N.J. Krogan, A. Emili, J.F. Greenblatt, L. Harrington, D. Lydall, and D. Durocher, "A Genome-Wide Screen Identifies the Evolutionarily Conserved KEOPS Complex as a Telomere Regulator", Cell, vol. 124, pp. 1155-1168, 2006. http://dx.doi.org/10.1016/j.cell.2005.12.044
- E. Kisseleva-Romanova, R. Lopreiato, A. Baudin-Baillieu, J. Rousselle, L. Ilan, K. Hofmann, A. Namane, C. Mann, and D. Libri, "Yeast homolog of a cancer-testis antigen defines a new transcription complex", The EMBO Journal, vol. 25, pp. 3576-3585, 2006. http://dx.doi.org/10.1038/sj.emboj.7601235
- A. Hecker, M. Graille, E. Madec, D. Gadelle, E. Le Cam, H. van Tilbergh, and P. Forterre, "The universal Kae1 protein and the associated Bud32 kinase (PRPK), a mysterious protein couple probably essential for genome maintenance in Archaea and Eukarya", Biochemical Society Transactions, vol. 37, pp. 29-35, 2009. http://dx.doi.org/10.1042/BST0370029
- J. Oberto, N. Breuil, A. Hecker, F. Farina, C. Brochier-Armanet, E. Culetto, and P. Forterre, "Qri7/OSGEPL, the mitochondrial version of the universal Kae1/YgjD protein, is essential for mitochondrial genome maintenance", Nucleic Acids Research, vol. 37, pp. 5343-5352, 2009. http://dx.doi.org/10.1093/nar/gkp557
- C.A. Lin, S.R. Ellis, and H.L. True, "The Sua5 Protein Is Essential for Normal Translational Regulation in Yeast", Molecular and Cellular Biology, vol. 30, pp. 354-363, 2010. http://dx.doi.org/10.1128/MCB.00754-09
- M. Hampsey, J. Na, I. Pinto, D. Ware, and R. Berroteran, "Extragenic suppressors of a translation initiation defect in the cyc1 gene of Saccharomyces cerevisiae", Biochimie, vol. 73, pp. 1445-1455, 1991. http://dx.doi.org/10.1016/0300-9084(91)90177-3
- K. Natarajan, M.R. Meyer, B.M. Jackson, D. Slade, C. Roberts, A.G. Hinnebusch, and M.J. Marton, "Transcriptional Profiling Shows that Gcn4p Is a Master Regulator of Gene Expression during Amino Acid Starvation in Yeast", Molecular and Cellular Biology, vol. 21, pp. 4347-4368, 2001. http://dx.doi.org/10.1128/MCB.21.13.4347-4368.2001
- M. Foiani, A.M. Cigan, C.J. Paddon, S. Harashima, and A.G. Hinnebusch, "GCD2, a translational repressor of the GCN4 gene, has a general function in the initiation of protein synthesis in Saccharomyces cerevisiae.", Molecular and Cellular Biology, vol. 11, pp. 3203-3216, 1991. http://dx.doi.org/10.1128/MCB.11.6.3203
- A.G. Hinnebusch, "TRANSLATIONAL REGULATION OFGCN4AND THE GENERAL AMINO ACID CONTROL OF YEAST", Annual Review of Microbiology, vol. 59, pp. 407-450, 2005. http://dx.doi.org/10.1146/annurev.micro.59.031805.133833
- M. Daugeron, T.L. Lenstra, M. Frizzarin, B. El Yacoubi, X. Liu, A. Baudin-Baillieu, P. Lijnzaad, L. Decourty, C. Saveanu, A. Jacquier, F.C.P. Holstege, V. de Crécy-Lagard, H. van Tilbeurgh, and D. Libri, "Gcn4 misregulation reveals a direct role for the evolutionary conserved EKC/KEOPS in the t6A modification of tRNAs", Nucleic Acids Research, vol. 39, pp. 6148-6160, 2011. http://dx.doi.org/10.1093/nar/gkr178
- M.P. Guy, and E.M. Phizicky, "Conservation of an intricate circuit for crucial modifications of the tRNAPhe anticodon loop in eukaryotes", RNA, vol. 21, pp. 61-74, 2014. http://dx.doi.org/10.1261/rna.047639.114
- M.P. Guy, B.M. Podyma, M.A. Preston, H.H. Shaheen, K.L. Krivos, P.A. Limbach, A.K. Hopper, and E.M. Phizicky, "Yeast Trm7 interacts with distinct proteins for critical modifications of the tRNAPhe anticodon loop", RNA, vol. 18, pp. 1921-1933, 2012. http://dx.doi.org/10.1261/rna.035287.112
- A. Noma, Y. Kirino, Y. Ikeuchi, and T. Suzuki, "Biosynthesis of wybutosine, a hyper-modified nucleoside in eukaryotic phenylalanine tRNA", The EMBO Journal, vol. 25, pp. 2142-2154, 2006. http://dx.doi.org/10.1038/sj.emboj.7601105
-
P.C. Thiaville, B. El Yacoubi, C. Köhrer, J.J. Thiaville, C. Deutsch, D. Iwata‐Reuyl, J.M. Bacusmo, J. Armengaud, Y. Bessho, C. Wetzel, X. Cao, P.A. Limbach, U.L. RajBhandary, and V. de Crécy‐Lagard, "Essentiality of threonylcarbamoyladenosine (t6
A ), a universal tRNA modification, in bacteria", Molecular Microbiology, vol. 98, pp. 1199-1221, 2015. http://dx.doi.org/10.1111/mmi.13209 - T. Taniguchi, K. Miyauchi, D. Nakane, M. Miyata, A. Muto, S. Nishimura, and T. Suzuki, "Decoding system for the AUA codon by tRNA Ile with the UAU anticodon in Mycoplasma mobile", Nucleic Acids Research, vol. 41, pp. 2621-2631, 2013. http://dx.doi.org/10.1093/nar/gks1344
- M.A. Machnicka, K. Milanowska, O. Osman Oglou, E. Purta, M. Kurkowska, A. Olchowik, W. Januszewski, S. Kalinowski, S. Dunin-Horkawicz, K.M. Rother, M. Helm, J.M. Bujnicki, and H. Grosjean, "MODOMICS: a database of RNA modification pathways—2013 update", Nucleic Acids Research, vol. 41, pp. D262-D267, 2012. http://dx.doi.org/10.1093/nar/gks1007
- G.R. Björk, B. Huang, O.P. Persson, and A.S. Byström, "A conserved modified wobble nucleoside (mcm5s2U) in lysyl-tRNA is required for viability in yeast", RNA, vol. 13, pp. 1245-1255, 2007. http://dx.doi.org/10.1261/rna.558707
- M. Dewez, F. Bauer, M. Dieu, M. Raes, J. Vandenhaute, and D. Hermand, "The conserved Wobble uridine tRNA thiolase Ctu1–Ctu2 is required to maintain genome integrity", Proceedings of the National Academy of Sciences, vol. 105, pp. 5459-5464, 2008. http://dx.doi.org/10.1073/pnas.0709404105
- M.J.O. Johansson, A. Esberg, B. Huang, G.R. Björk, and A.S. Byström, "Eukaryotic Wobble Uridine Modifications Promote a Functionally Redundant Decoding System", Molecular and Cellular Biology, vol. 28, pp. 3301-3312, 2008. http://dx.doi.org/10.1128/MCB.01542-07
- G. Jäger, K. Nilsson, and G.R. Björk, "The Phenotype of Many Independently Isolated +1 Frameshift Suppressor Mutants Supports a Pivotal Role of the P-Site in Reading Frame Maintenance", PLoS ONE, vol. 8, pp. e60246, 2013. http://dx.doi.org/10.1371/journal.pone.0060246
- J. URBONAVIC̆IUS, G. STAHL, J.M. DURAND, S.N. BEN SALEM, Q. QIAN, P.J. FARABAUGH, and G.R. BJÖRK, "Transfer RNA modifications that alter +1 frameshifting in general fail to affect −1 frameshifting", RNA, vol. 9, pp. 760-768, 2003. http://dx.doi.org/10.1261/rna.5210803
- B. HUANG, M.J. JOHANSSON, and A.S. BYSTRÖM, "An early step in wobble uridine tRNA modification requires the Elongator complex", RNA, vol. 11, pp. 424-436, 2005. http://dx.doi.org/10.1261/rna.7247705
- C. Chen, B. Huang, M. Eliasson, P. Rydén, and A.S. Byström, "Elongator Complex Influences Telomeric Gene Silencing and DNA Damage Response by Its Role in Wobble Uridine tRNA Modification", PLoS Genetics, vol. 7, pp. e1002258, 2011. http://dx.doi.org/10.1371/journal.pgen.1002258
- F. Bauer, and D. Hermand, "A coordinated codon-dependent regulation of translation by Elongator", Cell Cycle, vol. 11, pp. 4524-4529, 2012. http://dx.doi.org/10.4161/cc.22689
- A. Esberg, B. Huang, M.J. Johansson, and A.S. Byström, "Elevated Levels of Two tRNA Species Bypass the Requirement for Elongator Complex in Transcription and Exocytosis", Molecular Cell, vol. 24, pp. 139-148, 2006. http://dx.doi.org/10.1016/j.molcel.2006.07.031
- B. Zinshteyn, and W.V. Gilbert, "Loss of a Conserved tRNA Anticodon Modification Perturbs Cellular Signaling", PLoS Genetics, vol. 9, pp. e1003675, 2013. http://dx.doi.org/10.1371/journal.pgen.1003675
- D. Rojas-Benítez, C. Ibar, and �. Glavic, "TheDrosophilaEKC/KEOPS complex", Fly, vol. 7, pp. 168-172, 2013. http://dx.doi.org/10.4161/fly.25227
- D. Rojas-Benitez, P.C. Thiaville, V. de Crécy-Lagard, and A. Glavic, "The Levels of a Universally Conserved tRNA Modification Regulate Cell Growth", Journal of Biological Chemistry, vol. 290, pp. 18699-18707, 2015. http://dx.doi.org/10.1074/jbc.M115.665406
- C. Ibar, V.F. Cataldo, C. Vásquez-Doorman, P. Olguín, and �. Glavic, "Drosophilap53-related protein kinase is required for PI3K/TOR pathway-dependent growth", Development, vol. 140, pp. 1282-1291, 2013. http://dx.doi.org/10.1242/dev.086918
- V. Scheidt, . , A. Juedes, C. Baer, R. Klassen, R. Schaffrath, and . , "Loss of wobble uridine modification in tRNA anticodons interferes with TOR pathway signaling", Microbial Cell, vol. 1, pp. 416-424, 2014. http://dx.doi.org/10.15698/mic2014.12.179
- P. Thiaville, . , V. de Crecy-Lagard, . , . , and . , "The emerging role of complex modifications of tRNALysUUU in signaling pathways", Microbial Cell, vol. 2, pp. 1-4, 2015. http://dx.doi.org/10.15698/mic2015.01.185
- D. Nedialkova, and S. Leidel, "Optimization of Codon Translation Rates via tRNA Modifications Maintains Proteome Integrity", Cell, vol. 161, pp. 1606-1618, 2015. http://dx.doi.org/10.1016/j.cell.2015.05.022
- A. Naor, P.C. Thiaville, N. Altman-Price, I. Cohen-Or, T. Allers, V. de Crécy-Lagard, and U. Gophna, "A Genetic Investigation of the KEOPS Complex in Halophilic Archaea", PLoS ONE, vol. 7, pp. e43013, 2012. http://dx.doi.org/10.1371/journal.pone.0043013
- K. Kuranda, V. Leberre, S. Sokol, G. Palamarczyk, and J. François, "Investigating the caffeine effects in the yeast Saccharomyces cerevisiae brings new insights into the connection between TOR, PKC and Ras/cAMP signalling pathways", Molecular Microbiology, vol. 61, pp. 1147-1166, 2006. http://dx.doi.org/10.1111/j.1365-2958.2006.05300.x
- F. Alings, L.P. Sarin, C. Fufezan, H.C. Drexler, and S.A. Leidel, "An evolutionary approach uncovers a diverse response of tRNA 2-thiolation to elevated temperatures in yeast", RNA, vol. 21, pp. 202-212, 2014. http://dx.doi.org/10.1261/rna.048199.114
- R. Klassen, P. Grunewald, K.L. Thüring, C. Eichler, M. Helm, and R. Schaffrath, "Loss of Anticodon Wobble Uridine Modifications Affects tRNALys Function and Protein Levels in Saccharomyces cerevisiae", PLOS ONE, vol. 10, pp. e0119261, 2015. http://dx.doi.org/10.1371/journal.pone.0119261
- J.M. Kingsbury, and J.H. McCusker, "Homoserine Toxicity in Saccharomyces cerevisiae and Candida albicans Homoserine Kinase ( thr1 Δ) Mutants", Eukaryotic Cell, vol. 9, pp. 717-728, 2010. http://dx.doi.org/10.1128/EC.00044-10
- C. Katz, I. Cohen-Or, U. Gophna, and E.Z. Ron, "The Ubiquitous Conserved Glycopeptidase Gcp Prevents Accumulation of Toxic Glycated Proteins", mBio, vol. 1, 2010. http://dx.doi.org/10.1128/mBio.00195-10
- C. Tsai, and C.E. Frasch, "A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels", Analytical Biochemistry, vol. 119, pp. 115-119, 1982. http://dx.doi.org/10.1016/0003-2697(82)90673-X
- J. Anderson, L. Phan, R. Cuesta, B.A. Carlson, M. Pak, K. Asano, G.R. Björk, M. Tamame, and A.G. Hinnebusch, "The essential Gcd10p–Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA", Genes & Development, vol. 12, pp. 3650-3662, 1998. http://dx.doi.org/10.1101/gad.12.23.3650
- R. Balakrishnan, J. Park, K. Karra, B.C. Hitz, G. Binkley, E.L. Hong, J. Sullivan, G. Micklem, and J. Michael Cherry, "YeastMine—an integrated data warehouse for Saccharomyces cerevisiae data as a multipurpose tool-kit", Database, vol. 2012, 2012. http://dx.doi.org/10.1093/database/bar062
- A. Nishimura, T. Kotani, Y. Sasano, and H. Takagi, "An antioxidative mechanism mediated by the yeast N-acetyltransferase Mpr1: oxidative stress-induced arginine synthesis and its physiological role", FEMS Yeast Research, vol. 10, pp. 687-698, 2010. http://dx.doi.org/10.1111/j.1567-1364.2010.00650.x
- C.T. Harbison, D.B. Gordon, T.I. Lee, N.J. Rinaldi, K.D. Macisaac, T.W. Danford, N.M. Hannett, J. Tagne, D.B. Reynolds, J. Yoo, E.G. Jennings, J. Zeitlinger, D.K. Pokholok, M. Kellis, P.A. Rolfe, K.T. Takusagawa, E.S. Lander, D.K. Gifford, E. Fraenkel, and R.A. Young, "Transcriptional regulatory code of a eukaryotic genome", Nature, vol. 431, pp. 99-104, 2004. http://dx.doi.org/10.1038/nature02800
- S.L. Wolin, and P. Walter, "Ribosome pausing and stacking during translation of a eukaryotic mRNA.", The EMBO journal, 1988. http://www.ncbi.nlm.nih.gov/pubmed/2850168
- R. Legendre, A. Baudin-Baillieu, I. Hatin, and O. Namy, "RiboTools: a Galaxy toolbox for qualitative ribosome profiling analysis", Bioinformatics, vol. 31, pp. 2586-2588, 2015. http://dx.doi.org/10.1093/bioinformatics/btv174
- C.E. Pearson, "Repeat Associated Non-ATG Translation Initiation: One DNA, Two Transcripts, Seven Reading Frames, Potentially Nine Toxic Entities!", PLoS Genetics, vol. 7, pp. e1002018, 2011. http://dx.doi.org/10.1371/journal.pgen.1002018
- S. Chen, G. Lin, K. Chang, L. Yeh, and C. Wang, "Translational Efficiency of a Non-AUG Initiation Codon Is Significantly Affected by Its Sequence Context in Yeast", Journal of Biological Chemistry, vol. 283, pp. 3173-3180, 2008. http://dx.doi.org/10.1074/jbc.M706968200
- J.M. Clements, T.M. Laz, and F. Sherman, "Efficiency of translation initiation by non-AUG codons in Saccharomyces cerevisiae.", Molecular and Cellular Biology, vol. 8, pp. 4533-4536, 1988. http://dx.doi.org/10.1128/MCB.8.10.4533
- C. Chang, S. Chen, C. Lin, T. Wang, and C. Wang, "A single sequence context cannot satisfy all non-AUG initiator codons in yeast†", BMC Microbiology, vol. 10, 2010. http://dx.doi.org/10.1186/1471-2180-10-188
- K. Chang, and C. Wang, "Translation Initiation from a Naturally Occurring Non-AUG Codon in Saccharomyces cerevisiae", Journal of Biological Chemistry, vol. 279, pp. 13778-13785, 2004. http://dx.doi.org/10.1074/jbc.M311269200
- H. Tang, L. Yeh, N. Chen, T. Ripmaster, P. Schimmel, and C. Wang, "Translation of a Yeast Mitochondrial tRNA Synthetase Initiated at Redundant non-AUG Codons", Journal of Biological Chemistry, vol. 279, pp. 49656-49663, 2004. http://dx.doi.org/10.1074/jbc.M408081200
- P.M. Sharp, and W. Li, "The codon adaptation index-a measure of directional synonymous codon usage bias, and its potential applications", Nucleic Acids Research, vol. 15, pp. 1281-1295, 1987. http://dx.doi.org/10.1093/nar/15.3.1281
- M.D. Reis, "Solving the riddle of codon usage preferences: a test for translational selection", Nucleic Acids Research, vol. 32, pp. 5036-5044, 2004. http://dx.doi.org/10.1093/nar/gkh834
- S. Pechmann, and J. Frydman, "Evolutionary conservation of codon optimality reveals hidden signatures of cotranslational folding", Nature Structural & Molecular Biology, vol. 20, pp. 237-243, 2012. http://dx.doi.org/10.1038/nsmb.2466
- M. Stadler, and A. Fire, "Wobble base-pairing slows in vivo translation elongation in metazoans", RNA, vol. 17, pp. 2063-2073, 2011. http://dx.doi.org/10.1261/rna.02890211
- J. Gardin, R. Yeasmin, A. Yurovsky, Y. Cai, S. Skiena, and B. Futcher, "Measurement of average decoding rates of the 61 sense codons in vivo", eLife, vol. 3, 2014. http://dx.doi.org/10.7554/eLife.03735
- E.B. Kramer, and P.J. Farabaugh, "The frequency of translational misreading errors in E. coli is largely determined by tRNA competition", RNA, vol. 13, pp. 87-96, 2006. http://dx.doi.org/10.1261/rna.294907
- P.J. Farabaugh, "How translational accuracy influences reading frame maintenance", The EMBO Journal, vol. 18, pp. 1427-1434, 1999. http://dx.doi.org/10.1093/emboj/18.6.1427
- K. Kawakami, S. Pande, B. Faiola, D.P. Moore, J.D. Boeke, P.J. Farabaugh, J.N. Strathern, Y. Nakamura, and D.J. Garfinkel, "A rare tRNA-Arg(CCU) that regulates Ty1 element ribosomal frameshifting is essential for Ty1 retrotransposition in Saccharomyces cerevisiae.", Genetics, 1993. http://www.ncbi.nlm.nih.gov/pubmed/8243996
- N. Manickam, N. Nag, A. Abbasi, K. Patel, and P.J. Farabaugh, "Studies of translational misreading in vivo show that the ribosome very efficiently discriminates against most potential errors", RNA, vol. 20, pp. 9-15, 2013. http://dx.doi.org/10.1261/rna.039792.113
- S.K. DUBE, K.A. MARCKER, B.F.C. CLARK, and S. CORY, "Nucleotide Sequence of N-Formyl-methionyl-transfer RNA", Nature, vol. 218, pp. 232-233, 1968. http://dx.doi.org/10.1038/218232a0
- J.H. Miller, "GUG and UUG are initiation codons in vivo", Cell, vol. 1, pp. 73-76, 1974. http://dx.doi.org/10.1016/0092-8674(74)90063-4
- J.G. Files, K. Weber, C. Coulondre, and J.H. Miller, "Identification of the UUG codon as a translational initiation codon in vivo", Journal of Molecular Biology, vol. 95, pp. 327-330, 1975. http://dx.doi.org/10.1016/0022-2836(75)90398-8
- J.W. Stewart, F. Sherman, N.A. Shipman, and M. Jackson, "Identification and mutational relocation of the AUG codon initiating translation of iso-1-cytochrome c in yeast.", The Journal of biological chemistry, 1971. http://www.ncbi.nlm.nih.gov/pubmed/5135310
- F.E. BARALLE, and G.G. BROWNLEE, "AUG is the only recognisable signal sequence in the 5′ non-coding regions of eukaryotic mRNA", Nature, vol. 274, pp. 84-87, 1978. http://dx.doi.org/10.1038/274084a0
- J. WEISSENBACH, and H. GROSJEAN, "Effect of Threonylcarbamoyl Modification (t6A) in Yeast tRNAArgIII on Codon‐Anticodon and Anticodon‐Anticodon Interactions", European Journal of Biochemistry, vol. 116, pp. 207-213, 1981. http://dx.doi.org/10.1111/j.1432-1033.1981.tb05320.x
- J.M. Zaborske, V.L. Bauer DuMont, E.W.J. Wallace, T. Pan, C.F. Aquadro, and D.A. Drummond, "A Nutrient-Driven tRNA Modification Alters Translational Fidelity and Genome-wide Protein Coding across an Animal Genus", PLoS Biology, vol. 12, pp. e1002015, 2014. http://dx.doi.org/10.1371/journal.pbio.1002015
- T. Ikemura, "Correlation between the abundance of yeast transfer RNAs and the occurrence of the respective codons in protein genes", Journal of Molecular Biology, vol. 158, pp. 573-597, 1982. http://dx.doi.org/10.1016/0022-2836(82)90250-9
- L. Kraakman, K. Lemaire, P. Ma, A.W. Teunissen, M.C. Donaton, P. Van Dijck, J. Winderickx, J.H. De Winde, and J.M. Thevelein, "A Saccharomyces cerevisiae G‐protein coupled receptor, Gpr1, is specifically required for glucose activation of the cAMP pathway during the transition to growth on glucose", Molecular Microbiology, vol. 32, pp. 1002-1012, 1999. http://dx.doi.org/10.1046/j.1365-2958.1999.01413.x
- J.M. Cherry, E.L. Hong, C. Amundsen, R. Balakrishnan, G. Binkley, E.T. Chan, K.R. Christie, M.C. Costanzo, S.S. Dwight, S.R. Engel, D.G. Fisk, J.E. Hirschman, B.C. Hitz, K. Karra, C.J. Krieger, S.R. Miyasato, R.S. Nash, J. Park, M.S. Skrzypek, M. Simison, S. Weng, and E.D. Wong, "Saccharomyces Genome Database: the genomics resource of budding yeast", Nucleic Acids Research, vol. 40, pp. D700-D705, 2011. http://dx.doi.org/10.1093/nar/gkr1029
- M. Kato, and F.J. Slack, "Ageing and the small, non-coding RNA world", Ageing Research Reviews, vol. 12, pp. 429-435, 2013. http://dx.doi.org/10.1016/j.arr.2012.03.012
- J.G. Na, I. Pinto, and M. Hampsey, "Isolation and characterization of SUA5, a novel gene required for normal growth in Saccharomyces cerevisiae.", Genetics, 1992. http://www.ncbi.nlm.nih.gov/pubmed/1325384
- J.P. Miller, Z. Hussain, and M.P. Schweizer, "The involvement of the anticodon adjacent modified nucleoside N-[9-( -D-ribofuranosyl) purine-6-ylcarbamoyl]-threonine in the biological function of E.coli tRNAile", Nucleic Acids Research, vol. 3, pp. 1185-1202, 1976. http://dx.doi.org/10.1093/nar/3.5.1185
- R. Loewith, and M.N. Hall, "Target of Rapamycin (TOR) in Nutrient Signaling and Growth Control", Genetics, vol. 189, pp. 1177-1201, 2011. http://dx.doi.org/10.1534/genetics.111.133363
- J.R. Rohde, R. Bastidas, R. Puria, and M.E. Cardenas, "Nutritional control via Tor signaling in Saccharomyces cerevisiae", Current Opinion in Microbiology, vol. 11, pp. 153-160, 2008. http://dx.doi.org/10.1016/j.mib.2008.02.013
- T.W. Turowski, I. Karkusiewicz, J. Kowal, and M. Boguta, "Maf1-mediated repression of RNA polymerase III transcription inhibits tRNA degradation via RTD pathway", RNA, vol. 18, pp. 1823-1832, 2012. http://dx.doi.org/10.1261/rna.033597.112
- M.E. Cardenas, N.S. Cutler, M.C. Lorenz, C.J. Di Como, and J. Heitman, "The TOR signaling cascade regulates gene expression in response to nutrients", Genes & Development, vol. 13, pp. 3271-3279, 1999. http://dx.doi.org/10.1101/gad.13.24.3271
- P.J. Farabaugh, "PROGRAMMED TRANSLATIONAL FRAMESHIFTING", Annual Review of Genetics, vol. 30, pp. 507-528, 1996. http://dx.doi.org/10.1146/annurev.genet.30.1.507
- S. Tumu, A. Patil, W. Towns, M. Dyavaiah, and T.J. Begley, "The gene-specific codon counting database: a genome-based catalog of one-, two-, three-, four- and five-codon combinations present in Saccharomyces cerevisiae genes", Database, vol. 2012, 2012. http://dx.doi.org/10.1093/database/bas002
- E.T. Powers, and W.E. Balch, "Costly Mistakes: Translational Infidelity and Protein Homeostasis", Cell, vol. 134, pp. 204-206, 2008. http://dx.doi.org/10.1016/j.cell.2008.07.005
- K.J. Travers, C.K. Patil, L. Wodicka, D.J. Lockhart, J.S. Weissman, and P. Walter, "Functional and Genomic Analyses Reveal an Essential Coordination between the Unfolded Protein Response and ER-Associated Degradation", Cell, vol. 101, pp. 249-258, 2000. http://dx.doi.org/10.1016/s0092-8674(00)80835-1
- B. Herzog, B. Popova, A. Jakobshagen, H. Shahpasandzadeh, and G.H. Braus, "Mutual Cross Talk between the Regulators Hac1 of the Unfolded Protein Response and Gcn4 of the General Amino Acid Control of Saccharomyces cerevisiae", Eukaryotic Cell, vol. 12, pp. 1142-1154, 2013. http://dx.doi.org/10.1128/EC.00123-13
- R.D. Gietz, and R.H. Schiestl, "Frozen competent yeast cells that can be transformed with high efficiency using the LiAc/SS carrier DNA/PEG method", Nature Protocols, vol. 2, pp. 1-4, 2007. http://dx.doi.org/10.1038/nprot.2007.17
- S.C. Pomerantz, and J.A. McCloskey, "[44] Analysis of RNA hydrolyzates by liquid chromatography-mass spectrometry", Mass Spectrometry, pp. 796-824, 1990. http://dx.doi.org/10.1016/0076-6879(90)93452-q
- A. Baudin-Baillieu, R. Legendre, C. Kuchly, I. Hatin, S. Demais, C. Mestdagh, D. Gautheret, and O. Namy, "Genome-wide Translational Changes Induced by the Prion [PSI+]", Cell Reports, vol. 8, pp. 439-448, 2014. http://dx.doi.org/10.1016/j.celrep.2014.06.036
- A. Baudin-Baillieu, I. Hatin, R. Legendre, and O. Namy, "Translation Analysis at the Genome Scale by Ribosome Profiling", Methods in Molecular Biology, pp. 105-124, 2016. http://dx.doi.org/10.1007/978-1-4939-3079-1_7
- M. Martin, "Cutadapt removes adapter sequences from high-throughput sequencing reads", EMBnet.journal, vol. 17, pp. 10, 2011. http://dx.doi.org/10.14806/ej.17.1.200
- B. Langmead, and S.L. Salzberg, "Fast gapped-read alignment with Bowtie 2", Nature Methods, vol. 9, pp. 357-359, 2012. http://dx.doi.org/10.1038/nmeth.1923
- S.R. Engel, F.S. Dietrich, D.G. Fisk, G. Binkley, R. Balakrishnan, M.C. Costanzo, S.S. Dwight, B.C. Hitz, K. Karra, R.S. Nash, S. Weng, E.D. Wong, P. Lloyd, M.S. Skrzypek, S.R. Miyasato, M. Simison, and J.M. Cherry, "The Reference Genome Sequence ofSaccharomyces cerevisiae: Then and Now", G3 Genes|Genomes|Genetics, vol. 4, pp. 389-398, 2014. http://dx.doi.org/10.1534/g3.113.008995
- S. Anders, and W. Huber, "Differential expression analysis for sequence count data", Genome Biology, vol. 11, 2010. http://dx.doi.org/10.1186/gb-2010-11-10-r106
- S. Anders, "Analysing RNA-Seq data with the DESeq package", 1–28., 2012.
- Y. Benjamini, and Y. Hochberg, "Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing", J R Stat Soc Ser B 57(1): 289–300., 1995.
- V.V. Kushnirov, "Rapid and reliable protein extraction from yeast.", Yeast (Chichester, England), 2000. http://www.ncbi.nlm.nih.gov/pubmed/10861908
- A. Koplin, S. Preissler, Y. Ilina, M. Koch, A. Scior, M. Erhardt, and E. Deuerling, "A dual function for chaperones SSB–RAC and the NAC nascent polypeptide–associated complex on ribosomes", Journal of Cell Biology, vol. 189, pp. 57-68, 2010. http://dx.doi.org/10.1083/jcb.200910074
SUPPLEMENTAL INFORMATION
![]() Download Supplemental Information
Download Supplemental Information
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (grant number R01 GM70641 to V.dC-L.), by FONDECYT 1140522 and FONDAP 15090007 grants to A.G., and CONICYT grant AT24121519 to D.R.B. P.C.T. was funded in part by a Chateaubriand Fellowship from the French Embassy in the United States. RL was supported by a grant from ANR N° 11-BSV6-011-01 to ON. G.C. was funded by the Medical Research Council and the Gates Cambridge Scholarship. We thank Alan Hinnebush and Sebastian Leidel for plasmids and advice. The authors acknowledge Alex Moskalenko from the University of Florida Research Computing (http://researchcomputing.ufl.edu) for providing computational support that have contributed to the research results reported in this publication. The authors wish to acknowledge Henri Grosjean for his storied career in tRNA biology and modification. He has provided all of us with immeasurable inspiration and advice, and we wish him a happy retirement.
COPYRIGHT
© 2015

Global translational impacts of the loss of the tRNA modification t6A in yeast by Patrick C. Thiaville is licensed under a Creative Commons Attribution 4.0 International License.