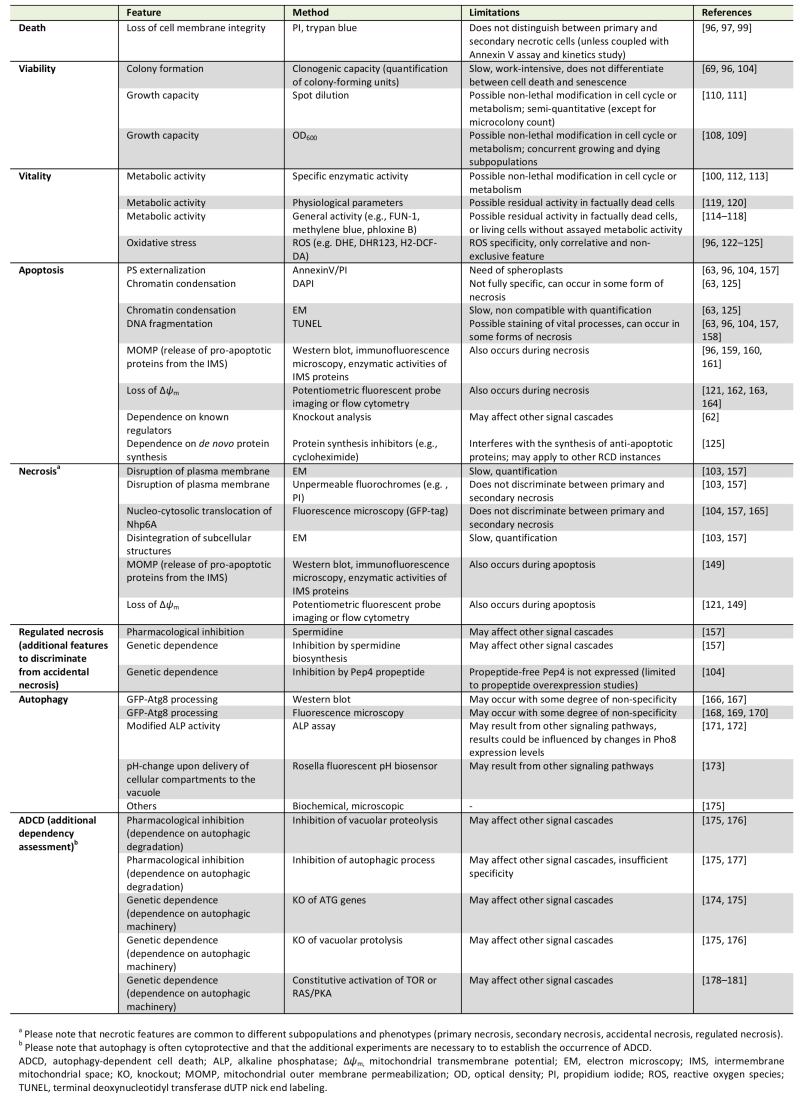
Back to article: Guidelines and recommendations on yeast cell death nomenclature
62. Carmona-Gutierrez D, Eisenberg T, Büttner S, Meisinger C, Kroemer G, and Madeo F (2002). Apoptosis in yeast: triggers, pathways, subroutines. Cell Death Differ 17(5): 763–773. http://dx.doi.org/10.1038/cdd.2009.219
63. Madeo F, Fröhlich E, and Fröhlich KU (2003). A yeast mutant showing diagnostic markers of early and late apoptosis. J Cell Biol 139(3): 729–734. https://www.ncbi.nlm.nih.gov/pubmed/?term=9348289
69. Madeo F, Herker E, Maldener C, Wissing S, Lächelt S, Herlan M, Fehr M, Lauber K, Sigrist SJ, Wesselborg S, and Fröhlich KU (2002). A caspase-related protease regulates apoptosis in yeast. Mol Cell 9(4): 911–917. https://www.ncbi.nlm.nih.gov/pubmed/?term=11983181
96. Büttner S, Eisenberg T, Carmona-Gutierrez D, Ruli D, Knauer H, Ruckenstuhl C, Sigrist C, Wissing S, Kollroser M, Fröhlich K-U, Sigrist S, and Madeo F (2007). Endonuclease G regulates budding yeast life and death. Mol Cell 25(2): 233–246. http://dx.doi.org/10.1016/j.molcel.2006.12.021
97. Deere D, Shen J, Vesey G, Bell P, Bissinger P, and Veal D (1998). Flow cytometry and cell sorting for yeast viability assessment and cell selection. Yeast 14(2): 147–160. http://dx.doi.org/10.1111/j.1749-6632.2009.04682.x
100. Kwolek-Mirek M, and Zadrag-Tecza R (2014). Comparison of methods used for assessing the viability and vitality of yeast cells. FEMS Yeast Res 14(7): 1068–1079. http://dx.doi.org/10.1111/1567-1364.12202
103. Eisenberg T, Carmona-Gutierrez D, Büttner S, Tavernarakis N, and Madeo F (2010). Necrosis in yeast. Apoptosis 15(3): 257–268. http://dx.doi.org/10.1007/s10495-009-0453-4
104. Carmona-Gutiérrez D, Bauer MA, Ring J, Knauer H, Eisenberg T, Büttner S, Ruckenstuhl C, Reisenbichler A, Magnes C, Rechberger GN, Birner-Gruenberger R, Jungwirth H, Fröhlich K-U, Sinner F, Kroemer G, and Madeo F (2011). The propeptide of yeast cathepsin D inhibits programmed necrosis. Cell Death Dis 2(5): e161. http://dx.doi.org/10.1038/cddis.2011.43
108. Jung PP, Christian N, Kay DP, Skupin A, and Linster CL (2015). Protocols and programs for high-throughput growth and aging phenotyping in yeast. PLoS One 10(3): e0119807. http://dx.doi.org/10.1371/journal.pone.0119807
109. Powers RW, Kaeberlein M, Caldwell SD, Kennedy BK, and Fields S (2006). Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev 20(2): 174–184. http://dx.doi.org/10.1101/gad.1381406
110. Giorgini F, Guidetti P, Nguyen Q, Bennett SC, and Muchowski PJ (2005). A genomic screen in yeast implicates kynurenine 3-monooxygenase as a therapeutic target for Huntington disease. Nat Genet 37(5): 526–531. http://dx.doi.org/10.1038/ng1542
111. Teng X, and Hardwick JM (2013). Quantification of Genetically Controlled Cell Death in Budding Yeast. Methods Mol Biol 1004: 161–170. http://dx.doi.org/10.1007/978-1-62703-383-1_12
112. Breeuwer P, Drocourt JL, Bunschoten N, Zwietering MH, Rombouts FM, and Abee T (1995). Characterization of uptake and hydrolysis of fluorescein diacetate and carboxyfluorescein diacetate by intracellular esterases in Saccharomyces cerevisiae, which result in accumulation of fluorescent product. Appl Environ Microbiol 61(4): 1614–1619. https://www.ncbi.nlm.nih.gov/pubmed/?term=7747975
113. Czekanska EM (2011). Assessment of cell proliferation with resazurin-based fluorescent dye. Methods Mol Biol 740: 27–32. http://dx.doi.org/10.1007/978-1-61779-108-6_5
114. Fannjiang Y, Cheng W-C, Lee SJ, Qi B, Pevsner J, McCaffery JM, Hill RB, Basañez G, and Hardwick JM (2004). Mitochondrial fission proteins regulate programmed cell death in yeast. Genes Dev 18(22): 2785–2797. http://dx.doi.org/10.1101/gad.1247904
115. Teng X, and Hardwick JM (2009). Reliable method for detection of programmed cell death in yeast. Methods Mol Biol 559: 335–342. http://dx.doi.org/10.1007/978-1-60327-017-5_23
116. Kuhn DM, Balkis M, Chandra J, Mukherjee PK, and Ghannoum MA (2003). Uses and Limitations of the XTT Assay in Studies of Candida Growth and Metabolism. J Clin Microbiol 41(1): 506–508. http://dx.doi.org/10.1128/JCM.41.1.506-508.2003
117. Bapat P, Nandy SK, Wangikar P, and Venkatesh KV (2006). Quantification of metabolically active biomass using Methylene Blue dye Reduction Test (MBRT): Measurement of CFU in about 200 s . J Microbiol Methods 65(1): 107–116. http://dx.doi.org/10.1016/j.mimet.2005.06.010
118. Noda T (2008). Viability Assays to Monitor Yeast Autophagy. In: Methods Enzymol, editor DJ Klionsky. Academic Press pp 27–32. http://dx.doi.org/10.1016/S0076-6879(08)03202-3
119. Anséhn S, and Nilsson L (1984). Direct membrane-damaging effect of ketoconazole and tioconazole on Candida albicans demonstrated by bioluminescent assay of ATP. Antimicrob Agents Chemother 26(1): 22–25. http://dx.doi.org/10.1128/AAC.26.1.22
120. Ludovico P, Sansonetty F, and Côrte-Real M (2001). Assessment of mitochondrial membrane potential in yeast cell populations by flow cytometry. Microbiol Read Engl 147(Pt 12): 3335–3343. http://dx.doi.org/10.1099/00221287-147-12-3335
121. Cottet-Rousselle C, Ronot X, Leverve X, and Mayol JF (2011). Cytometric assessment of mitochondria using fluorescent probes. Cytometry A Cytometry A. http://dx.doi.org/10.1002/cyto.a.21061
122. Perrone GG, Tan S-X, and Dawes IW (2008). Reactive oxygen species and yeast apoptosis. Biochim Biophys Acta 1783(7): 1354–1368. http://dx.doi.org/10.1016/j.bbamcr.2008.01.023
123. Lam YT, Aung-Htut MT, Lim YL, Yang H, and Dawes IW (2011). Changes in reactive oxygen species begin early during replicative aging of Saccharomyces cerevisiae cells. v 50(8): 963–970. http://dx.doi.org/10.1016/j.freeradbiomed.2011.01.013
124. Laun P, Pichova A, Madeo F, Fuchs J, Ellinger A, Kohlwein S, Dawes I, Fröhlich KU, and Breitenbach M (2001). Aged mother cells of Saccharomyces cerevisiae show markers of oxidative stress and apoptosis. Mol Microbiol 39(5): 1166–1173. https://www.ncbi.nlm.nih.gov/pubmed/?term=11251834
125. Madeo F, Fröhlich E, Ligr M, Grey M, Sigrist SJ, Wolf DH, and Fröhlich KU (1999). Oxygen stress: a regulator of apoptosis in yeast. J Cell Biol 145(4): 757–767. https://www.ncbi.nlm.nih.gov/pubmed/?term=10330404
149. Eastwood MD, and Meneghini MD (2015). Developmental Coordination of Gamete Differentiation with Programmed Cell Death in Sporulating Yeast. Eukaryot Cell 14(9): 858–867. http://dx.doi.org/10.1128/EC.00068-15
157. Eisenberg T, Knauer H, Schauer A, Büttner S, Ruckenstuhl C, Carmona-Gutierrez D, Ring J, Schroeder S, Magnes C, Antonacci L, Fussi H, Deszcz L, Hartl R, Schraml E, Criollo A, Megalou E, Weiskopf D, Laun P, Heeren G, Breitenbach M, Grubeck-Loebenstein B, Herker E, Fahrenkrog B, Fröhlich K-U, Sinner F, Tavernarakis N, Minois N, Kroemer G, and Madeo F (2009. Induction of autophagy by spermidine promotes longevity. Nat Cell Biol 11(11): 1305–1314. http://dx.doi.org/10.1038/ncb1975
158. Singh K, Kang PJ, and Park H-O (2008). The Rho5 GTPase is necessary for oxidant-induced cell death in budding yeast. Proc Natl Acad Sci U S A 105(5): 1522–1527. http://dx.doi.org/10.1073/pnas.0707359105
159. Ludovico P, Rodrigues F, Almeida A, Silva MT, Barrientos A, and Côrte-Real M (2002). Cytochrome c release and mitochondria involvement in programmed cell death induced by acetic acid in Saccharomyces cerevisiae. Mol Biol Cell 13(8): 2598–2606. http://dx.doi.org/10.1091/mbc.E01-12-0161
160. Wissing S, Ludovico P, Herker E, Büttner S, Engelhardt SM, Decker T, Link A, Proksch A, Rodrigues F, Corte-Real M, Fröhlich K-U, Manns J, Candé C, Sigrist SJ, Kroemer G, and Madeo F (2004). An AIF orthologue regulates apoptosis in yeast. J Cell Biol 166(7): 969–974. http://dx.doi.org/10.1083/jcb.200404138
161. Pérez-Gallardo RV, Briones LS, Díaz-Pérez AL, Gutiérrez S, Rodríguez-Zavala JS, and Campos-García J (2013). Reactive oxygen species production induced by ethanol in Saccharomyces cerevisiae increases because of a dysfunctional mitochondrial iron-sulfur cluster assembly system. FEMS Yeast Res 13(8): 804–819. http://dx.doi.org/: 10.1111/1567-1364.12090
162. Gourlay CW, and Ayscough KR (2005). Identification of an upstream regulatory pathway controlling actin-mediated apoptosis in yeast. J Cell Sci 118(10): 2119–2132. http://dx.doi.org/10.1242/jcs.02337
163. Pringle JR, Preston RA, Adams AE, Stearns T, Drubin DG, Haarer BK, and Jones EW (1989). Fluorescence microscopy methods for yeast. Methods Cell Biol 31: 357–435. https://www.ncbi.nlm.nih.gov/pubmed/?term=2476649
164. Hughes AL, and Gottschling DE (2012). An early age increase in vacuolar pH limits mitochondrial function and lifespan in yeast. Nature 492(7428): 261–265. http://dx.doi.org/10.1038/nature11654
165. Rockenfeller P, Ring J, Muschett V, Beranek A, Buettner S, Carmona-Gutierrez D, Eisenberg T, Khoury C, Rechberger G, Kohlwein SD, Kroemer G, and Madeo F (2010). Fatty acids trigger mitochondrion-dependent necrosis. Cell Cycle 9(14): 2836–2842. https://www.ncbi.nlm.nih.gov/pubmed/?term=20647757
166. Cheong H, Yorimitsu T, Reggiori F, Legakis JE, Wang C-W, and Klionsky DJ (2005). Atg17 regulates the magnitude of the autophagic response. Mol Biol Cell 16(7): 3438–3453. http://dx.doi.org/10.1091/mbc.E04-10-0894
167. Shintani T, and Klionsky DJ (2004). Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway.. J Biol Chem 279(29): 29889–29894. http://dx.doi.org/10.1074/jbc.M404399200
168. Cheong H, and Klionsky DJ (2008). Biochemical methods to monitor autophagy-related processes in yeast. Methods Enzymol 451: 1–26. http://dx.doi.org/10.1016/S0076-6879(08)03201-1
169. Kim J, Huang WP, and Klionsky DJ (2001 J Cell Biol 152(1): 51–64. https://www.ncbi.nlm.nih.gov/pubmed/?term=11149920
170. Shintani T, and Reggiori F (2008). Fluorescence microscopy-based assays for monitoring yeast Atg protein trafficking. Methods Enzymol 451: 43–56. http://dx.doi.org/10.1016/S0076-6879(08)03204-7
171. Camougrand N, Kissová I, Salin B, and Devenish RJ (2008). Monitoring mitophagy in yeast. Methods Enzymol 451: 89–107. http://dx.doi.org/10.1016/S0076-6879(08)03208-4
172. Noda T, and Klionsky DJ (2008). The quantitative Pho8Delta60 assay of nonspecific autophagy. Methods Enzymol 451: 33–42. http://dx.doi.org/10.1016/S0076-6879(08)03203-5
173. Rosado CJ, Mijaljica D, Hatzinisiriou I, Prescott M, and Devenish RJ (2008). Rosella: a fluorescent pH-biosensor for reporting vacuolar turnover of cytosol and organelles in yeast. Autophagy 4(2): 205–213. https://www.ncbi.nlm.nih.gov/pubmed/?term=18094608
174. Sampaio-Marques B, Felgueiras C, Silva A, Rodrigues M, Tenreiro S, Franssens V, Reichert AS, Outeiro TF, Winderickx J, and Ludovico P (2012). SNCA (α-synuclein)-induced toxicity in yeast cells is dependent on sirtuin 2 (Sir2)-mediated mitophagy. Autophagy 8(10): 1494–1509. http://dx.doi.org/10.4161/auto.21275
175. Klionsky DJ et al. (2016). Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12(1): 1–222. http://dx.doi.org/10.1080/15548627.2015.1100356
176. Delorme-Axford E, Guimaraes RS, Reggiori F, and Klionsky DJ (2015). The yeast Saccharomyces cerevisiae: an overview of methods to study autophagy progression. Methods 75: 3–12. http://dx.doi.org/10.1016/j.ymeth.2014.12.008
177. Yang Y, Hu L, Zheng H, Mao C, Hu W, Xiong K, Wang F, and Liu C (2013). Application and interpretation of current autophagy inhibitors and activators. Acta Pharmacol Sin 34(5): 625–635. http://dx.doi.org/10.1038/aps.2013.5
178. Binda M, Péli-Gulli M-P, Bonfils G, Panchaud N, Urban J, Sturgill TW, Loewith R, and De Virgilio C (2009). The Vam6 GEF controls TORC1 by activating the EGO complex. Mol Cell 35(5): 563–573. http://dx.doi.org/10.1016/j.molcel.2009.06.033
179. Budovskaya YV, Stephan JS, Reggiori F, Klionsky DJ, and Herman PK (2004). The Ras/cAMP-dependent protein kinase signaling pathway regulates an early step of the autophagy process in Saccharomyces cerevisiae. J Biol Chem 279(20): 20663–20671. http://dx.doi.org/10.1074/jbc.M400272200
180. Kira S, Tabata K, Shirahama-Noda K, Nozoe A, Yoshimori T, and Noda T (2014). Reciprocal conversion of Gtr1 and Gtr2 nucleotide-binding states by Npr2-Npr3 inactivates TORC1 and induces autophagy. Autophagy 10(9): 1565–1578. http://dx.doi.org/10.4161/auto.29397
181. Yorimitsu T, Zaman S, Broach JR, and Klionsky DJ (2007). Protein kinase A and Sch9 cooperatively regulate induction of autophagy in Saccharomyces cerevisiae. Mol Biol Cell 18(10): 4180–4189. http://dx.doi.org/10.1091/mbc.E07-05-0485