Reviews:
Microbial Cell, Vol. 5, No. 1, pp. 4 - 31; doi: 10.15698/mic2018.01.607
Guidelines and recommendations on yeast cell death nomenclature
1 Institute of Molecular Biosciences, NAWI Graz, University of Graz, Graz, Austria. 2 Centro Andaluz de Biología, Molecular y Medicina Regenerativa-CABIMER, Universidad de Sevilla, Sevilla, Spain. 3 Department of Biology, Providence College, Providence, USA. 4 Department of Biomedical Science, University of Sheffield, Sheffield, United Kingdom. 5 Department of Physiology and Biochemistry, University of Malta, Msida, Malta. 6 Department of Biochemistry and Molecular Biology, George S. Wise Faculty of Life Sciences, Tel Aviv University, Tel Aviv, Israel. 7 Department of Biochemistry and Molecular Biology, University of Miami Miller School of Medicine, Miami, USA. 8 Department of Neurology, University of Miami Miller School of Medicine, Miami, USA. 9 Department of Molecular Microbiology and Immunology, Brown University, Providence, USA. 10 Institut National de la Santé et de la Recherche Médicale UMR1078, Université de Bretagne Occidentale, Etablissement Français du Sang Bretagne, CHRU Brest, Hôpital Morvan, Laboratoire de Génétique Moléculaire, Brest, France. 11 Institute of Cell Biology, University of Bayreuth, Bayreuth, Germany. 12 Department of Cell Biology and Physiology, Division of Genetics, University of Salzburg, Salzburg, Austria. 13 Department of Molecular and Cellular Biology, Roswell Park Cancer Institute, Buffalo, NY, USA. 14 Department of Molecular Biosciences, The Wenner-Gren Institute, Stockholm University, Stockholm, Sweden. 15 Department of Biology, University of Florence, Firenze, Italy. 16 European Research Institute for the Biology of Ageing, University of Groningen, University Medical Center Groningen, Groningen, The Netherlands. 17 Dept. Molecular Biology, Graduate School of Biomedical Sciences, Rowan University, Stratford, USA. 18 Center of Molecular and Environmental Biology, Department of Biology, University of Minho, Braga, Portugal. 19 Instituto de Investigação e Inovação em Saúde, Universidade do Porto, Porto, Portugal. 20 Instituto de Biologia Molecular e Celular, Universidade do Porto, Porto, Portugal. 21 Departamento de Biologia Molecular, Instituto de Ciências Biomédicas Abel Salazar, Universidade do Porto, Porto, Portugal. 22 CNRS, University of Bordeaux CBMN (UMR 5248), Pessac, France. 23 School of Biotechnology and Biomolecular Sciences, University of New South Wales, Sydney, Australia. 24 Department of Biology, University of Fribourg, Fribourg, Switzerland. 25 Institute for Plant Genomics and Biotechnology, Texas A&M University, Texas, USA. 26 BioTechMed Graz, Graz, Austria. 27 Laboratory Biology of the Nucleus, Institute for Molecular Biology and Medicine, Université Libre de Bruxelles, Charleroi, Belgium. 28 Department of Biochemistry, University of Lausanne, Lausanne, Switzerland. 29 Laboratoire de Biotechnologie Moléculaire des Eucaryotes, Center de Biotechnologie de Sfax, Sfax, Tunisia. 30 Institute of Biomembranes, Bioenergetics and Molecular Biotechnologies, National Research Council, Bari, Italy. 31 Department of Chemistry, Life Sciences and Environmental Sustainability, University of Parma, Parma, Italy. 32 Kent Fungal Group, School of Biosciences, University of Kent, Canterbury, United Kingdom. 33 Faculty of Biology, Medicine and Health, The University of Manchester, Manchester, United Kingdom. 34 Department of Chemistry and Chemical Engineering, Royal Military College, Kingston, Ontario, Canada. 35 Zürich, Switzerland. 36 Department of Biology and Chemistry, University of Osnabrück, Osnabrück, Germany. 37 Heinrich Pette Institute, Leibniz Institute for Experimental Virology, Hamburg, Germany. 38 Cell Biology, University of Kaiserslautern, Kaiserslautern, Germany. 39 Department of Systems Biology, University of Alcalá, Alcalá de Henares, Spain. 40 Division of Internal Medicine, The University of Texas MD Anderson Cancer Center, Houston, Texas, USA. 41 Life and Health Sciences Research Institute (ICVS), School of Health Sciences, University of Minho, Minho, Portugal. 42 ICVS/3B’s – PT Government Associate Laboratory, Braga/Guimarães, Portugal. 43 Institut de Biochimie et de Génétique Cellulaires, UMR5095, CNRS & Université de Bordeaux, Bordeaux, France. 44 Department of Biotechnolgy and Biosciences, University of Milano-Bicocca, Milano, Italy. 45 Instituto Pasteur-Fondazione Cenci Bolognetti – Department of Biology and Biotechnology “C. Darwin”, La Sapienza University of Rome, Rome, Italy. 46 Sprott Center for Stem Cell Research, Ottawa Hospital Research Institute, The Ottawa Hospital, Ottawa, Canada. 47 Department of Cellular and Molecular Medicine, University of Ottawa, Ottawa, Canada. 48 Department of Medicine, Division of Cardiology, University of Ottawa, Ottawa, Canada. 49 Institute of Biochemistry and Molecular Biology, ZBMZ, Faculty of Medicine, University of Freiburg, Freiburg, Germany. 50 Department of Biology and Biological Engineering, Chalmers University of Technology, Gothenburg, Sweden. 51 Novo Nordisk Foundation Center for Biosustainability, Chalmers University of Technology, Gothenburg, Sweden. 52 Novo Nordisk Foundation Center for Biosustainability, Technical University of Denmark, DK2800 Lyngby, Denmark. 53 Institute for Biomedicine, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden. 54 Institute for Molecular Biosciences, Goethe University, Frankfurt am Main, Germany. 55 Department of Experimental Neurodegeneration, Center for Nanoscale Microscopy and Molecular Physiology of the Brain, Center for Biostructural Imaging of Neurodegeneration, University Medical Center Göttingen, Göttingen, Germany. 56 Max Planck Institute for Experimental Medicine, Göttingen, Germany. 57 Institute of Neuroscience, The Medical School, Newcastle University, Framlington Place, Newcastle Upon Tyne, NE2 4HH, United Kingdom. 58 CEDOC, Chronic Diseases Research Centre, NOVA Medical School, Faculdade de Ciências Médicas, Universidade NOVA de Lisboa, Lisboa, Portugal. 59 Department of Molecular Genetics, The Ohio State University, Columbus, OH, USA. 60 Malaria Research Unit, SMITh, ICBMS, UMR 5246 CNRS-INSA-CPE-University Lyon, Lyon, France. 61 Institut of Parasitology and Medical Mycology, Hospices Civils de Lyon, Lyon, France. 62 Department of Biochemistry, Faculty of Natural Sciences, Comenius University in Bratislava, Bratislava, Slovak Republic. 63 Department of Molecular and Cellular Biology, College of Biological Sciences, UC Davis, Davis, California, USA. 64 Biosciences, University of Exeter, Exeter, United Kingdom. 65 Department of Cell Biology and Physiology, Division of Genetics, University of Salzburg, Salzburg, Austria. 66 Institute of Biology, Division of Microbiology, University of Kassel, Kassel, Germany. 67 Department of Ecology, Faculty of Sciences, University of Malaga, Malaga, Spain. 68 A.N. Belozersky Institute of physico-chemical biology, Moscow State University, Moscow, Russia. 69 School of Plant Sciences and Food Security, Faculty of Life Sciences, Tel Aviv University, Tel Aviv, Israel. 70 Institute for Biology/Genetics, Freie Universität Berlin, Berlin, Germany. 71 Laboratory of Molecular Cell Biology, Institute of Botany and Microbiology, KU Leuven, Leuven, Belgium. 72 Center for Microbiology, VIB, Leuven-Heverlee, Belgium. 73 Centre of Microbial and Plant Genetics, KU Leuven, Leuven, Belgium. 74 Biology Department, Concordia University, Montreal, Canada. 75 Institute for Integrative Biology of the Cell (I2BC), SBIGEM, CEA-Saclay, Université Paris-Saclay, Gif-sur-Yvette, France. 76 Department of Biology, Functional Biology, KU Leuven, Leuven-Heverlee, Belgium. 77 Cevec Pharmaceuticals, Cologne, Germany. 78 Institute of Pharmacy and Molecular Biotechnology, Heidelberg University, Heidelberg, Germany. 79 Department of Zoology and Physiology, University of Wyoming, Laramie, USA. 80 Department of Pathology, University of Maryland School of Medicine, Baltimore, USA. 81 School of Life Sciences, Tsinghua University, Beijing, China. 82 Department of Radiation Oncology, Weill Cornell Medical College, New York, NY, USA. 83 Sandra and Edward Meyer Cancer Center, New York, NY, USA. 84 Université Paris Descartes/Paris V, Paris, France. 85 Equipe 11 Labellisée Ligue Contre le Cancer, Centre de Recherche des Cordeliers, Paris, France. 86 Cell Biology and Metabolomics Platforms, Gustave Roussy Comprehensive Cancer Center, Villejuif, France. 87 INSERM, U1138, Paris, France. 88 Université Pierre et Marie Curie/Paris VI, Paris, France. 89 Pôle de Biologie, Hôpital Européen Georges Pompidou, Paris, France. 90 Karolinska Institute, Department of Women’s and Children’s Health, Karolinska University Hospital, Stockholm, Sweden.
Keywords: accidental cell death, apoptosis, autophagic cell death, autophagy, caspases, mitochondrial membrane permeabilization, mitotic catastrophe, model organism, necrosis, reactive oxygen species, regulated cell death, Saccharomyces cerevisiae.
Abbreviations:
ACD - Accidental cell death,
ADCD - Autophagy-dependent cell death,
ALP - Alkaline phosphatase,
ATCD - Autophagic cell death,
ATG - Autophagy-related,
CFU - Colony-forming unit,
DAMP - Damage-associated molecular pattern,
DAPI - 4',6-diamidino-2-phenylindole,
DHE – Dihydroethidium,
DHR123 - Dihydrorhodamine 123,
EM - electron microscopy,
H2-DCF-DA - 2,7-dichlorodihydrofluorescein diacetate,
HMGB1 - High mobility group box 1,
IDAI - Indwelling device-associated infection,
IMS - Intermembrane mitochondrial space,
KO – Knockout, MC - Mitotic catastrophe,
MOMP - Mitochondrial outer membrane permeabilization,
MPT - Mitochondrial permeability transition,
NCCD - Nomenclature Committee on Cell Death,
OD - Optical density,
PCD - Programmed cell death,
PI - Propidium iodide, PS – Phosphatidylserine,
RCD - Regulated cell death,
ROS - Reactive oxygen species,
TUNEL - Terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling,
Δψm - Mitochondrial transmembrane potential.
Received originally: 19/12/2017 Accepted: 29/12/2017
Published: 01/01/2018
Correspondence:
Frank Madeo, Institute of Molecular Biosciences, University of Graz, Graz, Austria; frank.madeo@uni-graz.at
Didac Carmona-Gutierrez, Institute of Molecular Biosciences, University of Graz, Graz, Austria; carmonag@uni-graz.at
Guido Kroemer, INSERM, U1138, Paris, France kroemer@orange.fr
Lorenzo Galluzzi, Weill Cornell Medical College, New York, NY, USA deadoc@vodafone.it
Conflict of interest statement: The authors declare no conflict of interest.
Please cite this article as: Didac Carmona-Gutierrez, Maria Anna Bauer, Andreas Zimmermann, Andrés Aguilera, Nicanor Austriaco, Kathryn Ayscough, Rena Balzan, Shoshana Bar-Nun, Antonio Barrientos, Peter Belenky, Marc Blondel, Ralf J. Braun, Michael Breitenbach, William C. Burhans, Sabrina Büttner, Duccio Cavalieri, Michael Chang, Katrina F. Cooper, Manuela Côrte-Real, Vítor Costa, Christophe Cul-lin, Ian Dawes, Jörn Dengjel, Martin B. Dickman, Tobias Eisenberg, Birthe Fahrenkrog, Nicolas Fasel, Kai-Uwe Fröh-lich, Ali Gargouri, Sergio Giannattasio et al (2018). Guidelines and recommendations on yeast cell death nomencla-ture. Microbial Cell 5(1): 4-31. doi: 10.15698/mic2018.01.607
Abstract
Elucidating the biology of yeast in its full complexity has major implications for science, medicine and industry. One of the most critical processes determining yeast life and physiology is cellular demise. However, the investigation of yeast cell death is a relatively young field, and a widely accepted set of concepts and terms is still missing. Here, we propose unified criteria for the definition of accidental, regulated, and programmed forms of cell death in yeast based on a series of morphological and biochemical criteria. Specifically, we provide consensus guidelines on the differential definition of terms including apoptosis, regulated necrosis, and autophagic cell death, as we refer to additional cell death routines that are relevant for the biology of (at least some species of) yeast. As this area of investigation advances rapidly, changes and extensions to this set of recommendations will be implemented in the years to come. Nonetheless, we strongly encourage the authors, reviewers and editors of scientific articles to adopt these collective standards in order to establish an accurate framework for yeast cell death research and, ultimately, to accelerate the progress of this vibrant field of research.
INTRODUCTION
Yeast, a fungus that predominantly lives as a unicellular organism, has had an extraordinary influence on humanity throughout millennia, from its usage for baking and brewing to the potential of some species to cause life-threatening human diseases. The cultural, industrial, biotechnological, and medical impact of this organism remains unparalleled. The use of yeast fermentation to produce alcoholic beverages and to leaven bread coincided with the rise of ancient civilizations and has persisted until our days. Importantly, the continued development of yeast strains as vehicles for the development of new technology, for example in bioethanol, drug, and enzyme production, as well as the implementation of unconventional yeast species in industrial processes, highlights the ever increasing importance of yeast now and in the future [1][2]. This is exemplified by the fact that the global market for yeast products is in the multibillion dollar range and is expected to grow further [3]. Beyond the mentioned applications, yeast has a direct impact on human health and disease. Many fungi, including some yeasts, can exist as commensals, i.e., they are part of our natural microbiota, forming the mycobiome [4]. In fact, it is being increasingly recognized that fungi are a major determinant in establishing commensal microbial communities and are thus vital for healthy individuals [5]. However, under certain circumstances, e.g., compromised immunity, commensal fungi may become opportunistic pathogens and as such are a potential cause for infectious diseases [6]. These include superficial infections of the skin and nails (especially by dermatophytes) that affect billions worldwide, biofilm colonisations of mucosal surfaces and more serious invasive infections, which can have very high mortality rates and are estimated to lead to 1.5 million deaths per year [7]. A significant number of these deaths arise from infections caused by the yeasts Candida albicans, Candida glabrata and Cryptococcus neoformans in immunocompromised individuals. This socioeconomic burden is further amplified by the unprecedented rise in fungal diseases that are affecting plants and animals [8]. These examples highlight the importance of a full understanding of fungal biology, and the study of yeast cell biological processes has been crucial in this respect.
–
Yeasts have served as a successful research tool for the last century, Saccharomyces cerevisiae (the budding yeast) being one of the most thoroughly studied eukaryotes at the cellular and molecular levels. Indeed, yeast continues to be one of the preferred model organisms to explore eukaryotic cell biology, both due to its technical advantages in devising/sophisticating molecular tool kits to study cellular biology, and to a high degree of functional conservation [9]. Also, yeast offers rapid growth and inexpensive accessibility paired with a high amenability to biochemical and genetic manipulation. This enables the establishment of various experimental setups, ranging from single experiments to high-throughput, genome-scale, unbiased screenings in a short time frame. Notably, many insights obtained in yeast have proven to be transferable to higher eukaryotes. Indeed, over the past decades, yeast studies have unveiled individual gene functions as well as gene and protein interactions, and have instrumentally contributed to the understanding of fundamental cellular processes such as eukaryotic cell cycle control [10][11][12][13][14][15], autophagy [16][17][18][19], mitochondrial function [20][21], including mitochondrial import [22][23][24][25], protein degradation [26], vesicle fusion [27][28], genetic instability [29][30], epigenetic control [31][32], metabolic regulation [33][34][35], or cellular nutrient sensing [36].
–
In addition, studies on yeast have shed light on human diseases, providing a cellular platform to examine, for instance, prion biology, virus-host interactions, metabolic diseases, neurodegenerative disorders, cancer, or aging [37][38][39][40][41][42][43][44][45][46][47][48][49][50][51][52][53][54][55][56][57][58][59][60][61]. Among the pathophysiologically relevant pathways that can readily be explored in yeast are those governing cellular demise. Indeed, cell death regulation is structurally and functionally conserved in yeast [21][62][63][64][65][66], and yeast has even served to uncover and establish factors and pathways involved in apoptosis and other controlled cell death subroutines, which have later been corroborated in metazoan or other multicellular systems, e.g., the AAA-ATPase Cdc48/VCP [63] [67], the BAX inhibitor-1 [68], the implication of metacaspases as cell death regulators [69][70][71], the role of cathepsin D in non-autophagic mitochondrial degradation [72][73], or the lethal impact of ER-Golgi transport blockage as one of the mechanisms explaining the demise of dopaminergic neurons during Parkinson’s disease [74]. To sum up, on the one hand, cell death represents a key process that can be feasibly modeled in yeast. On the other hand, the understanding of yeast cell death and its putative modulation may improve industrial and biotechnological applications, provide insights into mycobiome dynamics, and help develop the fight against fungal and other diseases.
–
In multicellular organisms, the controlled suicide of single cells is crucial for development and homeostasis, providing a system that eliminates superfluous cells. The presence of such a mechanism also allows for the removal of damaged cells that might compromise organismal fitness. In a single-celled organism like yeast, this paradigm does not seem to apply at first sight, since – in this case – cellular suicide entails the death of the whole organism. However, in a way, a population of yeast cells de facto behave as a multicellular entity of communicating individuals rather than a group of isolated cells that do not interact with each other. In fact, a given yeast population originates from a single clone, and the ultimate biological goal of that population is the survival of the genetic information representing that very clone. Thus, under certain circumstances, the death of unfit or damaged yeast cells promotes the survival of the population as a whole. A number of physiological scenarios have been described that corroborate this teleological explanation for a cellular suicide program in yeast, including antagonistic interactions between yeasts, aging, mating, or colony formation [54][61][75][76][77][78][79][80][81][82][82][83][84][85]. Of note, also other unicellular organisms, including bacteria and protozoan parasites, incorporate regulatory processes that are at least partly reminiscent of higher eukaryotic cell death programs [86][87][88][89][90][91].
–
–
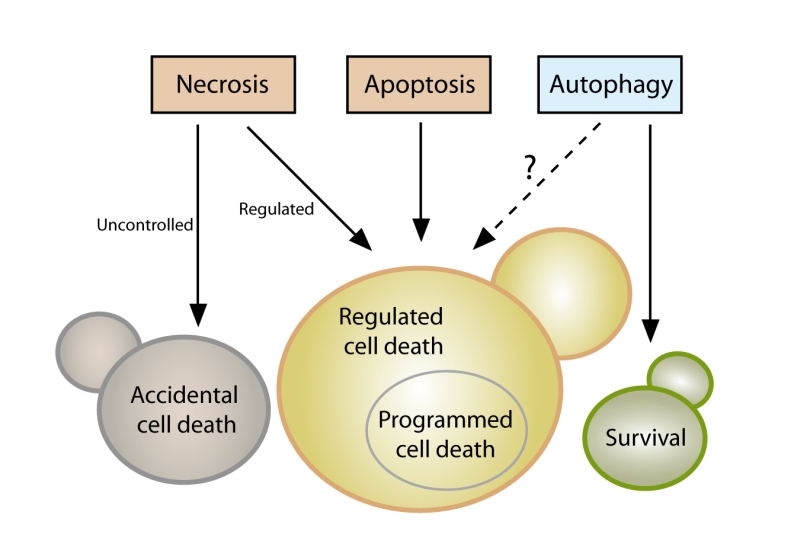
Even though it is now clear that yeast can indeed undergo cellular suicide, the corresponding terminology to describe this multifaceted process remains heterogenous and potentially misleading. Thus, we believe that there is timely need for a more precise and consistent nomenclature that clearly defines the concept of “yeast cell death”, considering morphological, enzymological, and functional aspects. Such standardization seems of importance, given that the field of yeast cell death is continuously expanding with significant progress being made at the phenotypical and mechanistic levels, including the finding that, akin to higher eukaryotes, yeast can also engage in distinct cell death modalities (Figure 1). In this paper we thus attempt for the first time to formulate a series of recommendations and caveats with respect to cell death-related results obtained in yeast. To this aim, we have followed the directions of the Nomenclature Committee on Cell Death (NCCD) [92][93][94][95] and adapted them to the particularities of Saccharomyces cerevisiae, which we think can be extended to other yeast species and to multicellular filamentous fungi. Our goal is to frame a uniform set of guidelines that facilitate the communication among yeast cell death researchers, ultimately supporting and accelerating scientific advance (Box 1). In that respect, the nomenclature presented herein will likely need to be revised and updated as the field of yeast cell death moves forward and even more precise definitions are required.
–
| describes cellular death following exposure to very harsh microenvironmental conditions. |
| represents a regulated cell death subroutine characterized by specific morphologic and biochemical features and executed via different pro-apoptotic factors; eventually, it culminates in secondary necrosis. |
| defines a predominantly cytoprotective process that orchestrates the digestion of intracellular material (e.g. proteins, organelles) in the vacuole. |
| describes a lethal subroutine, in which the molecular machinery of autophagy (or parts thereof) causally contributes to cellular demise. |
| defines a status of irreversible plasma membrane breakdown (only then, the cell is factually disintegrated, irrespectively of which upstream pathway or routine has been engaged). |
| reflects the ability of a cell to divide and thus to proliferate. |
| reflects the physiological capabilities of a cell and thus its metabolic activity. |
| is a cell death instance mainly characterized by plasma membrane permeabilization; primary necrosis (cellular necrosis occurring ab initio) may take place in an accidental or regulated manner; secondary necrosis (combined necrotic and apoptotic features) is the final stage of the apoptotic process. |
| designates a specific type of regulated cell death, which occurs in strictly physiological scenarios (e.g., development, aging). |
| describes cellular death occurring in the context of a failing response to internal or external mild stress. |
| is a regulated cell death modality with characteristic features of necrosis that can be inhibited by specific pharmacological or genetic interventions. |
YEAST CELL DEATH AND SURVIVAL
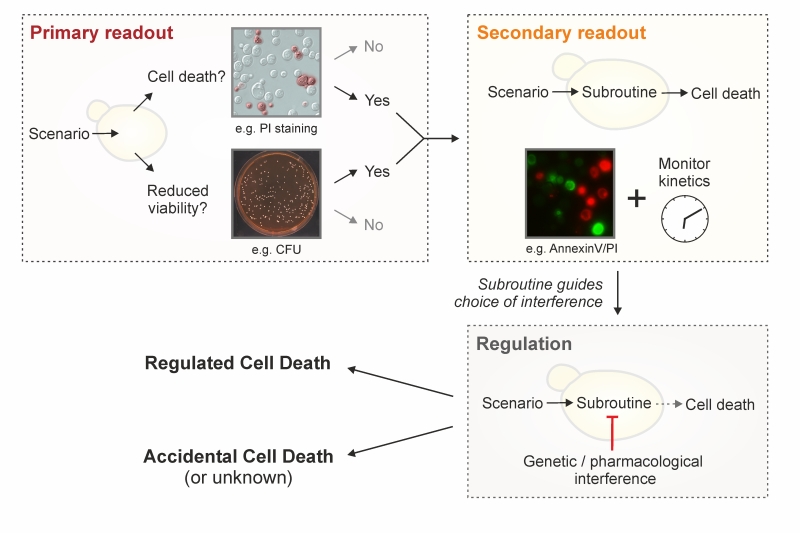
A crucial issue that demands a clear definition is the question of cell death itself. When is a cell dead? According to the NCCD guidelines this is only the case upon irreversible plasma membrane breakdown or complete cellular fragmentation, because only then the cell is factually disintegrated, irrespectively of which upstream pathway or routine has been engaged [93]. In fact, no earlier marker can be defined that reliably determines death in all settings. Thereby, this lethal irreversibility might start with the collapse of the electrochemical membrane potential across the plasma membrane through formation of a leak. In yeast, the most common method to monitor cell membrane integrity in vivo is to use propidium iodide (PI). PI is a fluorescent nucleic acid intercalator that can only enter cells with a ruptured cell membrane, and can be routinely employed in both low and high throughput formats [96][97][98]. Along similar lines, colorimetric dyes like trypan blue may be used, but are less common [99][100][101]. Further potential alternatives exist (e.g., 7-aminoactinomycin D), but will need to be thoroughly tested with respect to their suitability for yeast cell death applications in future studies. As mentioned, assessing cell membrane disintegrity is the only technique to quantify actual cell death and must be performed irrespectively of the lethal setting being analyzed. This is imperative, since lethal signaling does not imply that the final stage (cell death) is reached or even that it will be reached at a later stage (see below). In fact, specific subpopulations engaged in lethal pathways that maintain plasma membrane integrity (e.g., early apoptotic cells, see below) are by definition not (yet) dead. In that respect, timecourse experiments are important to monitor both the lethal subroutine-specific phenotypes and the actual occurrence of cell death over time. Of note, indications exist that upon specific stress insults, a small subpopulation of yeast retains the ability to repair cell membrane damage even after having stained positive for PI [102]. Given the lack of other comparably well established dyes in this context and the large body of data supporting PI staining as a valid method to quantify loss of survival, we conclude that determining PI positivity is – at this point – the best technique to quantitatively approach yeast cell death. Still, for the sake of accuracy and waiting for further evidence supporting the above-mentioned indications, we suggest expressing a corresponding quantification as “% PI-positivity” or “% cell death (PI positive)” instead of % death” or “% survival” upon using this method. In the long term, the development and establishment of alternative dyes should be explored in order to validate data obtained with PI. A number of approaches allow to experimentally assess (i) cell viability, which reflects the ability of a cell to divide, and (ii) cell vitality, defined as the physiological capabilities of a cell [100]. Nonetheless, an impaired/compromised (i) proliferation or (ii) metabolic capacity does not necessarily result in cellular demise. Thus, these techniques alone cannot be used to demonstrate cell death. Still, they are very useful to complement and corroborate data obtained with PI or alternative dyes.
–
Assessing clonogenicity with plating assays is the most commonly used method to quantify cell viability [62][103]. Here, a defined number of cells from a given culture are plated on rich medium agar plates that are further incubated to allow colony formation. The ratio between the resulting colony-forming units (CFUs) and the originally plated number of cells reflects the viability state in the culture. Theoretically, however, it is possible that under specific conditions (of genetic nature, for instance), colony formation may be blocked in cells that per se are still alive (a condition usually refered to as senescence). Additional caveats include the possibility that live cells at the point of plating might die before forming a colony and/or that the plating procedure itself might drive (a fraction of) cells into death, which would be indistinguishable from cell senescence. Nonetheless, the literature suggests clonogenic capacity as a very good correlate to cell death in a plethora of different settings [69][96][104][105] and thus represents a valid approximation to quantify survival in yeast populations. Of note, clonogenicity can also be measured by monitoring CFU formation at the microcolony level (time-lapse photomicroscopy) [106][107]. Even though cell and colony counting can be automated, clonogenicity assays are rather time-consuming and used for low- to medium-throughput analyses.
–
A further technique to assess yeast viability follows the growth rate of a given culture, which may decrease as a consequence of increased cell death. For this purpose, an aliquot is inoculated into fresh liquid medium and the growth is monitored, for instance, via photometric measurement of optical densities over a specific period of time [108][109]. Optionally, spot dilution assays can be performed, where cultures are spotted in serial dilutions on agar plates [110]. Here, the growth ability is compared between cultures at the various dilution steps in a semi-quantitative manner, although automated readout of microcolonies can be used to yield a quantitative result [111]. Monitoring growth can be scaled up and performed either manually or using robotics support, which makes it an attractive technique, especially for screen-based analyses. As with other viability assays, an important disadvantage is that a decreased growth rate can also result from a non-lethal event such as modulation of cell cycle progression or a reduced metabolism due to an alteration in the use of media components.
–
One possibility to evaluate yeast cell vitality is to directly assess the activity of specific enzymes directly. Although this is not widely employed in yeast cell death research, it represents an avenue to assay the physiological state of a metabolic pathway within the cell [100][112][113]. As pointed out below, a caveat of this approach is the possible distortion of results by residual activity in dead cells. A further option is to use vital dyes, like the two-color fluorescent probe FUN-1, which diffuses into cells, irrespectively of their viability status, and results in green fluorescence of the cytoplasm. Dead cells fluoresce green while (live) cells that have both plasma membrane integrity and metabolic capability, can further process the probe, resulting in red vacuolar fluorescence [114][115]. Similarly, several tetrazolium salts are reduced into colored formazan crystals [116]. Methylene blue is converted to the colorless leucomethylene blue only in metabolically active cells [117], while the red dye phloxine B is only retained in metabolically inactive cells that are unable to actively export it [100][118]. Other methods aim at assessing further aspects of cellular physiology, including the cellular ATP content (e.g., based on the luciferin-luciferase reaction) [119] or mitochondrial transmembrane potential (e.g., upon staining with rhodamine 123, JC-1, TMRM/E, DiOC6(3)) [120][121]. It should be noted that the readout of metabolic signatures has considerably improved with new generation extracellular flux analyzers, offering the possibility to simultaneously measure mitochondrial respiration and glycolysis (and thus mitochondrial function). A drawback of metabolic assays resides in the fact that cells may be able to maintain some metabolic activities until cell membrane rupture occurs, and that some rely on specific metabolic processes such as oxidative phosphorylation that are not mandatory for cell survival. Thus, such techniques may fail to detect subpopulations of dead (or alive) cells, reflecting the notion that a decrease in growth or metabolic activity (i.e., viability or vitality) cannot be placed on a par with an increase in cell death. In conclusion, as mentioned above, the term cell death should be used only upon observing breakdown of the plasma membrane and thus loss of cell integrity (e.g., upon PI staining). In addition, we suggest to strengthen this observation by simultaneously assessing clonogenic capacity (Figure 2), since (i) it represents the best-established output to accurately monitor overall cellular viability and (ii) it empirically correlates very strongly with actual cell death markers. Importantly, both methods are easy, quick and relatively inexpensive. The use of additional dyes/stainings/assays provides valuable complementary information, but cannot be used alone to unequivocally define a cell as dead.
–
–
Yeast cell death is often accompanied by oxidative damage and thus, a widely employed method in the field is the detection of reactive oxygen species (ROS) [122]. Indeed, a number of different ROS, like the superoxide anion, hydroxyl radical, and hydrogen peroxide, can accumulate upon mitochondrial disturbances, ER stress or other cellular derangements [96][122][123][124][125]. ROS can generally be detected using membrane-permeable dyes that are oxidized to fluorescent products in a ROS-dependent manner. Importantly, these stains do not measure ROS as a group, but rather react with specific species. For instance, dihydroethidium (DHE) preferentially reacts with superoxides, while dihydrorhodamine 123 (DHR123) and 2,7-dichlorodihydrofluorescein diacetate (H2-DCF-DA) are converted by a broad range of other ROS, but only poorly by superoxides [126]. Such specificities should be taken into account when measuring ROS with a particular stain, since distinct lethal triggers might result in the production of a differential ROS subset [123]. Thus, we recommend to specifically indicate the ROS subtype that is being monitored instead of generally referring to ROS production. Of note, to a certain degree, DHE may also be oxidized unspecifically (independently of superoxide). In order to exploit the full potential of DHE as a superoxide-specific dye, a range of methodological possibilities (e.g., the use of optimized spectra) exist [127][128]. The standardization of such refinements for DHE assays, which are a preferred tool in yeast cell death research, should be addressed in the future. While ROS measurements allow for high-throughput approaches due to their simplicity and relatively low cost, it is imperative to realize that this method does not discriminate between living and dead cells, although ROS usually precede and are often causative for cell death in yeast [125]. In fact, ROS play a crucial role in intracellular signaling [129][130][131][132], functioning, for instance, as direct and indirect regulators of diverse physiologically relevant targets [133][134][135]. In addition, limited ROS generation might be beneficial under certain conditions, since the resulting adaptive responses can promote stress resistance as a form of preconditioning (hormesis) [131][136][137][138][139]. Thus, an increase in ROS should be regarded as a cell death-correlated phenotype only in connection with assays that directly determine increased plasma membrane disintegration and loss of clonogenicity (see above). Similarly, a decrease in ROS production by incubation with anti-oxidants might support the mechanistic involvement of ROS in the lethal process, but only when cell death is adequately monitored.
ACCIDENTAL VERSUS REGULATED CELL DEATH
Cellular demise in yeast may occur in two mutually exclusive variants: either as an accidental event or through a regulated pathway. Accidental cell death (ACD) occurs upon exposure to severe conditions, resulting in a rapid, uncontrollable and unavoidable form of death. ACD may follow a series of extreme stimuli, including physical conditions, such as very high temperatures or pressures, severe chemical insults like strong detergents and high concentrations of acids or bases as well as mechanical challenges, for instance, vigorous shearing or ultrasonic treatment. The immediate nature of ACD, which is characterized by a virtually immediate structural breakdown of cells, allows no form of pharmacologic or genetic inhibition. Thus, this form of cell death does not constitute a direct target for modulation or prevention. However, it remains unclear whether yeast cells undergoing ACD may release endogenous, bioactive molecules to the extracellular space [75][79]. If so, such molecules could interact with local cells that have survived the primary insult and ignite a response within the whole yeast population. Such a consequence of ACD may resemble the release of damage-associated molecular patterns (DAMPs) by dying human cells. DAMPs can stimulate a direct or indirect (via innate immune effectors) cytotoxic response in surrounding bystander cells that have survived ACD [140][141][142][143][144]. In such a case, interfering with the effects of ACD on the rest of the population remains possible.
–
ACD is often equated with necrosis, which in yeast is usually identified as a cellular condition of early plasma membrane permeabilization in the absence of typical apoptotic markers and of complete disintegration of subcellular structures [103]. Indeed, ACD usually exhibits morphological features of necrosis, but mounting evidence suggests that – as it is the case in human cells – a physiologically relevant, regulated type of necrosis does also exist in yeast. Thus, we recommend to avoid using the term “necrosis” to define an accidental and uncontrollable type of death, and to favor the term “ACD”. We believe that this will avoid any potential misunderstandings regarding the two fundamentally dinstinct (accidental versus regulated) modalities of yeast cell death manifesting with a necrotic morphology (see below).
–
That said, many lethal stimuli result in a form of yeast cell death that – at odds with ACD – is executed by a genetically encoded, dedicated molecular machinery. In higher eukaryotes, a distinction is made between such a controlled form of cell death when it occurs (i) in the framework of a purely physiological program, e.g., during (post-) embryonic development or tissue homeostasis, or (ii) as a response to either a perturbation of intracellular or extracellular homeostasis, e.g., upon exposure to mild stress or as a consequence of mutations. Cell death occurring in the former scenario is termed “programmed cell death” (PCD), while the expression “regulated cell death” (RCD) encompasses both PCD as well as all other instances of cell death that depend on a molecular machinery [145][146][147][148].
–
For yeast cell death, many authors have used the term PCD to interchangeably refer to all types of cellular demise that are not accidental (i.e., to all instances of RCD). However, emerging evidence is confirming that a yeast population, be it a liquid culture or a solid colony, bears a degree of complexity reminiscent of multicellular organisms that demands a revision of this terminology. For instance, during yeast gametogenesis (or sporulation), immature meiotic products as well as the mother cell itself succumb via activation of vacuolar rupture [149][150]. Interestingly, the mother cell’s demise is delayed until spores have reached a threshold degree of differentiation. Thus, in this scenario, RCD occurs in the frame of a developmentally coordinated program, de facto representing an instance of PCD. During yeast chronological aging, the cellular community maintains homeostasis thanks to the programmed death of dysfunctional or old cells, which spares and provides nutrients to the fitter individuals [75][76]. In yeast colonies, stationary-phase or slow-growing cells differentiate into specific subpopulations with unique metabolic properties and particular functions within the colony [151][152]. These examples show that, indeed, yeast populations can harness cell death to control coordinated development, homeostasis and differentiation. Hence, we propose to define PCD in yeast as a specific instance of RCD that is executed in the frame of such physiologic programs. All other forms of regulated demise (e.g., cell death induction upon stress, or as a consequence of specific genetic alterations) should be referred to with the superordinate term of RCD.
–
Importantly, since RCD depends on a defined molecular machinery, it can be modulated with pharmacologic or genetic means. The extent of such modulation depends on the progression of the process across a hitherto poorly defined point-of-no-return. According to the NCCD, the processes preceding such point are part of cellular stress responses, while those following it belong to actual cell death signaling [93]. Adopting this rationale, RCD can be accelerated or delayed (but not avoided) if the point-of-no-return has been trespassed. Instead, prior to that point, modulating stress responses or avoiding stress can prevent RCD. However, the definition of this point-of-no-return has not been established yet, implying that the exact boundary between the reversibility of a stress stimulus and the irrevocable engagement in a lethal cascade remains to be specified.
–
| Besides the specific points to be addressed for appropriately classifying an observed cell death phenotype, various general issues need to be considered, as well. |
| (i) In general, we recommend to sequentially test the following: first, whether cell death occurs (defined as loss of plasma membrane integrity, which may be accompanied by decreased proliferation and/or diminished metabolic activity), second, the hallmarks of cell death subroutines as implied at the descriptive level (morphology, biochemistry), and third, the mechanisms of cell death as determined at the interventional level (genetic, pharmacological). Thus, it is imperative to combine multiple and complementary approaches, also with respect to kinetics (markers should preceed cell death), to characterize a specific cell death type. We recommend performing at least two independent and subroutine-specific assays, preferably not just restricted to the assessment of morphological features. |
| (ii) While mainly qualitative or arduously quantifiable methods (e.g. electron microscopy) offer the possibility to accurately define specific cell death phenotypes, they may be poorly representative of the general sample conditions. Thus, we encourage using these methods for exploratory purposes, but strongly suggest accompanying them with quantitative assays. |
| (iii) In many cell death settings, the dependence on specific factors may be tested by inhibiting their function. Where possible, we recommend employing genetic tools (i.e., knockout, temperature-sensitive mutants) instead of pharmacological inhibitors. Indeed, the specificity of such compounds might not be sufficient to precisely block the activity of a single pathway/factor that characterizes a cell death subroutine [153]. |
| (iv) If knockout strains are used to inquire the involvement of the corresponding gene/protein in a given cell death scenario, we recommend employing self-generated deletion strains and control results by complementation analysis (i.e., ectopic re-expression of detleted genes etc.). Those available at public strain collections constitute useful starting tools for experimentation, but may have accumulated secondary mutations that might lead to misinterpretations [154]. |
| (v) For the quantification of fluorescence-based detection methods, we recommend using flow cytometry rather than a fluorescent plate reader. Data obtained with a plate reader may indeed be influenced by the fluorescence of the entire culture, which may vary with several parameters including strain-specific cell size. Few highly fluorescent cells may yield the same signals compared to a substantial fraction of moderately or low fluorescent but still positively stained cells. Thus, even upon normalization to the OD600, bulk results are less accurate than results obtained with flow cytometry, which is based on actual single-cell fluorescence. Plate readers may be convenient for high-throughput studies, but positive hits should be validated using flow cytometry. |
–
Yeast RCD may follow different subroutines (see below) that can be differentiated from each other by a series of morphological and biochemical features. To precisely characterize the lethal phenotype, we recommend (i) to first determine if cell death actually occurs (as opposed to only reduced viability/vitality), (ii) if it does, to then examine the subroutine(s) involved via morphological and biochemical observations, using at least two different detection methods [155], and (iii) finally, to corroborate the implicated mechanism(s) via genetic and pharmacological interventions (Figure 2). Finally, it should be noted that in cell death research, it is generally advisable to determine the kinetics of the parameters under scrutiny [156]. In order to detect the differential appearance of apoptotic or necrotic characteristics, we recommend assessing such features at different time points to yield a better resolution of cell death events. Importantly, subroutine-specific markers should precede cell death. In the following sections, we will describe yeast RCD subroutines and the techniques to precisely discriminate amongst them (Table 1). Beyond the specificities outlined below, a number of general issues and notes of caution also need to be considered (Box 2).
–
Table 1. Methods commonly used for the assessment of cell death, viability and vitality as well as for the identification of different cell death subroutines in yeast. |
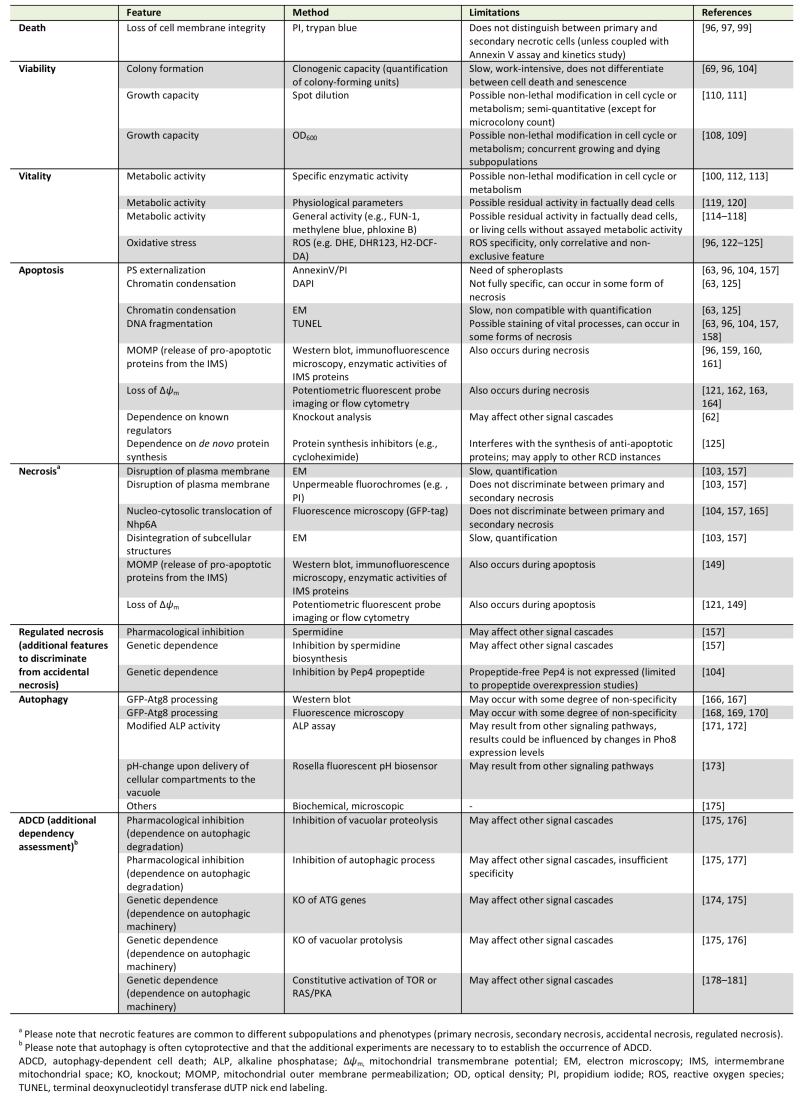 |
| [62] [63] [69] [96] [97] [99] [100] [103] [104] [108] [109] [110] [111] [112] [113][114] [115] [116] [117] [118] [119] [120] [121] [122][123] [124] [125] [149] [157] [158] [159] [160] [161] [162] [163] [164] [165] [166] [167] [168] [169] [170] [171] [172] [173] [174] [175] [176] [177] [178] [179] [180] [181] |
–
APOPTOSIS
Most studies on RCD in yeast have been conducted in the budding yeast Saccharomyces cerevisiae. This includes the first observation of an apoptotic phenotype in yeast, specifically in a strain with a point mutation in the gene coding for the cell cycle protein Cdc48 [63]. One of the early indications for an active cellular participation in the yeast apoptotic process was that RCD in this setting can be prevented by inhibiting de novo protein synthesis, e.g. by cycloheximide [125]. Ever since these discoveries, a set of methods has been established, validated and refined that allows to specifically determine whether a yeast cell has engaged in an apoptotic pathway [62]. These techniques are mainly based on the key morphologic and biochemical features of an apoptotic cell. We suggest employing at least two of these apoptosis-specific methods (one of them should be Annexin V staining, see below) and include at least one viability assay (preferably clonogenic capacity) to describe a corresponding phenotype.
–
One of the events most commonly associated with apoptosis is the exposure of phosphatidylserine (PS) on the outer leaflet of the plasma membrane [157]. However, PS externalization might be context-dependent to a certain degree, at least within the complexity of the human cellular network [93][158][159]. It remains unclear whether this is also the case in yeast, although the current evidence suggests that PS externalization is a universal feature of yeast cells undergoing apoptosis. PS externalization can be assessed via monitoring PS-binding to Annexin V, which is usually fluorescently labeled for quantitative (e.g., fluorescence reader-based or flow cytrometric analyses) and qualitative (microscopic) evaluation. To this aim, the cell wall needs to be (partially) digested in order to make the externalized PS accessible to Annexin V and permit binding. Usually, the Annexin V assay is performed as a co-staining with a marker for plasma membrane rupture like PI [63][96][104][160]. This allows for the discrimination between several subpopulations as they occur in yeast: (i) Annexin V/PI double negative, (ii) Annexin V positive, (iii) PI positive, and (iv) Annexin V/PI double positive cells.
–
We believe that the second (Annexin V positive) and third (PI positive) subpopulations can be readily interpreted as apoptotic and primary necrotic, respectively, provided that at least one more assay is performed to validate this assumption. For the fourth subpopulation (Annexin V/PI double positive cells), we favor the following interpretation: unlike multicellular animals, a yeast population presumably does not eliminate apoptotic cells via the phagocytic activity of other yeast cells. In the absence of such clearance by scavengers, an apoptotic cell eventually undergoes a metabolic collapse that results in breakdown of the plasma membrane integrity and thus a necrotic phenotype. This phenomenon is termed “secondary necrosis” to discriminate it from “primary necrosis”, which describes the phenotype of “cellular necrosis occurring ab initio” [161][162]. We thus view the above-mentioned fourth subgroup (Annexin V/PI double positive cells) as a late apoptotic population that has undergone secondary necrosis.
–
Still, these cells might also have undergone secondary necrosis following other cell death subroutines, but at this point there is no evidence for this possibility, which should be evaluated earlier in the cascade of events leading to cellular demise. Importantly, the phenotypical shift from apoptosis to secondary necrosis might reflect defined mo-ecular events and thus be experimentally distinguishable from ACD with necrotic features and primary necrosis also at the functional level [163][164]. It could be argued that, in turn, primary necrotic cells might eventually stain for apoptotic markers like Annexin V, thus also yielding AnnexinV/PI double-stained cells. However, necrotic markers do appear without apoptotic characteristics and such primary necrotic populations are stably maintained during long-term physiological conditions like chronological aging. This strongly suggests that primary necrosis can be distinguished from secondary necrosis by the absence or presence of apoptotic markers. Still, no study has yet systematically evaluated this distinction at the cellular level, for instance, via cell sorting analysis. Until such further analysis, this interpretation remains a valid approximation. In any case, we suggest determining the kinetics of the cell death process (see above) to accurately resolve the appearance of these subpopulations. In general, any approaches that facilitate monitoring death scenarios time-dependently represent a helpful improvement, for instance replicative age-associated changes using microfluidic platforms [165][166][167][168][169].
–
In multicellular animals, clearance of apoptotic cells is a central physiological feature for maintenance of organismal homeostasis. Still, secondary necrosis does occur under certain circumstances [162]. In vitro, cultured metazoan cells that are left to finalize the apoptotic process without interruption (e.g., without interference of phagocytic scavenging) eventually succumb with features of secondary necrosis [162][170][171]. In vivo, secondary necrosis may occur in multicellular animals, for example, when apoptotic cells are shed into the lumina of hollow organs with low probability to encounter scavengers or when apoptotic cell death occurs at a pace that surpasses the local scavenging capacity [162][172][173]. These observations suggest that secondary necrosis following apoptosis is a conserved outcome upon exposure to pro-apoptogenic stimuli if clearance mechanisms are absent or insufficient.
–
Besides PS externalization, apoptotic cells exhibit chromatin condensation, which can be readily assessed by nuclear staining with dies such as 4′,6-diamidino-2-phenylindole (DAPI) followed by microscopic inspection [63][125]. Another characteristic that accompanies yeast apoptosis – especially at late steps of the process – is DNA fragmentation. It is often assessed via the “terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling” (TUNEL) test, which allows for the fluorescent labelling of free 3′-hydroxyl ends that can be easily monitored via microscopy analysis and quantified using a fluorescent plate reader or a flow cytometer [63][96][104][160]. In many yeast cell death scenarios, TUNEL positivity matches apoptotic markers determined by other assays [96][104][160][174]. However, TUNEL staining detects free 3′-hydroxyl ends regardless of the molecular mechanism involved in generating them. In fact, in some conditions, necrosis, DNA repair, or active gene transcription have all been shown to yield TUNEL positivity, at least in human cells [175][176][177][178][179][180]. In yeast, the nature and the kinetics of DNA fragmentation detected by the TUNEL test need further investigation, even though previous studies have partly addressed these issues [79][181]. In summary, we recommend using the TUNEL test as a method to determine the occurrence of DNA fragmentation associated with yeast apoptosis rather than a technique for quantifying apoptosis on its own. In addition, the TUNEL test may provide an assay to screen for cellular demise in high-throughput assays. In this setting, hits must be confirmed by testing cellular membrane integrity and clonogenic capacity. Furthermore, apoptotic DNA damage may be tested using the so-called “comet assay”, or single cell gel electrophoresis, whereby physiologic DNA strand breaks are distinguished from apoptotic DNA dissolution in individual cells (the latter forms a distinct cluster of fragmented DNA at the ‘tail’ of the comet) [182]. In addition, the flow cytometric detection of a subpopulation with hypoploid DNA content (sub-G0/G1) has been previously employed as an alternative to assess apoptotic DNA degradation [183]. However, such results should be interpreted carefully, since apparent hypoploidy may also reflect an artefact from the debris associated with necrotic cells, unless discarded by cell sorting analyses [184].
–
Apoptotic cell death often follows mitochondrial outer membrane permeabilization (MOMP), which culminates with the release of pro-apoptotic proteins from the intermembrane space and irreversible loss of mitochondrial transmembrane potential (Δψm) [96][185][186][187][188][189][190][191]. A detailed analysis of these mitochondrial subevents requires precise kinetic determinations. For instance, in acetic-acid induced RCD, pro-apoptotic cytochrome c release, which depends on the ADP/ATP carrier [192], occurs before mitochondrial integrity is lost [193]. All of these biochemical features might be evaluated to determine an apoptotic phenotype, though it should be kept in mind that mitochondria have also been associated with at least one other RCD subroutine (regulated necrosis) [149]. Thus, we recommend the involvement of mitochondria in apoptosis to be validated by at least two specific methods (one of them should be assessing PS externalization) and at least one viability assay (preferentially clonogenic capacity).
–
A large number of apoptotic regulators and executors have been identified in yeast so far [62]. This enables testing whether RCD occurring upon a given stimulus is at least partly dependent on one of these factors based on genetic manipulations, pending confirmatory experiments with morphological and biochemical assays. We advise to interpret results from genetic disruption or inhibition studies with caution, as it is difficult to estimate whether other or how many signaling cascades have been affected by a manipulation a priori specific. Indeed, many yeast cell death regulators, e.g., cytochrome c, apoptosis-inducing factor (Aif1), endonuclease G (Nuc1) and the yeast metacaspase (Yca1), exert both lethal and vital functions [62][69][96][186][188][194][195][196][197]. Importantly, the molecular network underlying apoptosis regulation in yeast is starting to be uncovered and additional regulators and subroutines that are yet unknown are expected to emerge. Thus, if a given cell death phenotype is not dependent on any of the known apoptotic regulators this does not exclude apoptosis as a possible cell death modality.
–
For exploring a putative apoptotic mechanism in a given cell death scenario, the deletion strains of known apoptotic regulators should be harnessed, since distinct apoptotic subroutines exist that rely on different factors that may act independently from each other to orchestrate cellular demise. For instance, the yeast metacaspase Yca1 is involved in many apoptotic RCD and PCD settings [62][69][75]. Thus, cell death inhibition in yca1 knockout strains may point towards an apoptotic mechanism. However, under certain conditions, apoptosis is not executed via Yca1, but instead relies on other factors, including Aif1, Nuc1, the human cyclophilin D ortholog Cpr3, the BH3-only protein Ybh3 or ceramides [96][188][198][199][200][201][202][203][204]. Importantly, while yeast harbors a single metacaspase-encoding gene (YCA1), it is possible that other proteases might functionally substitute for metacaspases [205][206][207]. Thus, in cases where Yca1 is not involved in cell death regulation, we favor the expression “Yca1-independent” instead of “metacaspase-” or “caspase-independent” cell death. For cell death stimuli that are dependent on Yca1, we consider that the terms “Yca1-“, “metacaspase-“, and “caspase-dependent” are all appropriate. In fact, though much controversy has accompanied the denomination of metacaspases as true homologs of caspases, recent advances strongly indicate that this is the case [71]. Indeed, caspases and metacaspases seem to be evolutionary distinct variants with a functional commonality that do fulfill the criteria of homology, since they both share (i) a common cellular program (RCD) and (ii) common or at least overlapping substrates [70][208][209].
–
In human cells, extrinsic apoptosis defines a caspase-dependent cell death subroutine that is induced by extracellular lethal ligands. These ligands are sensed and transmitted either via specific transmembrane death receptors or through so-called ‘dependence receptors’. Dependence receptors can trigger two opposite signaling pathways: in the presence of ligand, they elicit signals involved in cell survival, migration and differentiation, but in the absence of ligand, they promote apoptotic RCD. Thus, dependence receptors only exert lethal functions when the concentration of their specific ligands falls below a critical threshold level [210]. While in yeast no such dedicated receptors are known, cases of metacaspase-dependent apoptosis induction by molecules that may operate from the extracellular microenvironment have been described. For instance, toxins secreted by virus-infected killer strains and a number of drugs have been shown to trigger apoptosis executed by Yca1 [82][211][212][213][214]. Yet, it remains unknown whether these factors act on intracellular targets, or whether they may also bind to plasma membrane-localized receptors. Given the complexity and interactivity of a yeast population, it is conceivable that a yet-to-be-determined extrinsic apoptotic pathway may co-regulate cell death within a yeast community [79][215]. However, and meeting the definition of extrinsic apoptosis put forward by the NCCD, we suggest not to use this term until dedicated death receptors or dependence receptors are discovered. Similarly, another specific type of apoptosis in human cells, anoikis, which defines a form of intrinsic apoptosis restricted to adherent cells that detach from the matrix [216], is theoretically possible in yeast. Indeed, adhesion mediated by cell-wall-bound adhesins is crucial for colony and biofilm formation as well as for host-pathogen interactions [217][218][219]. While it remains conceivable that normally adherent yeast cells, which detach in a specific scenario where adhesion is important, might undergo a form of anoikis, this form of RCD has not (yet) been described in yeast.
REGULATED NECROSIS
In dying yeast, necrotic characteristics may appear in the frame of a primary or secondary necrotic process. While secondary necrosis is probably a consequence of apoptosis in most if not all cases (see above), a primary necrotic phenotype (which occurs without any preceding apoptotic traits) may result from two cell death modalities: ACD or RCD. Indeed, yeast primary necrosis can not only be the outcome of severe insults (accidental necrosis), but also develop as an event orchestrated by a genetically controlled machinery (regulated necrosis) [103]. In both cases, cell death is characterized by a set of distinct morphological and biochemical features that defines it as necrotic.
–
Necrosis first leads to a gain in cell volume and organelle swelling (oncosis), which may be observed, for instance, using fluorescent microscopy of GFP-fused proteins that mark organellar membranes [149]. Eventually, necrotic cells also show the complete breakdown and disintegration of subcellular structures, which can be assessed using electron microscopy [160]. Similarly, the rupture of the plasma membrane that accompanies necrosis can easily be assayed via electron microscopy or fluorochromes like PI that only enter cells with a disintegrated cell membrane, but are excluded by healthy or early apoptotic cells [103][160]. In yeast, the release of intracellular material has not yet been systematically employed as an assay to characterize necrotic cell death. However, the nucleo-cytosolic translocation of Nhp6A may be used to assess necrosis in yeast [104][160][220][221]. Nhp6A is the yeast homolog of the mammalian protein high mobility group box 1 (HMGB1), whose release accompanies immunogenic cell death mammalian cells [144][222]. We suggest assessing at least two of these markers in order to define bona fide primary necrosis in yeast. In addition, viability should be measured with at least one assay (preferably by assessing clonogenic capacity) to corroborate cellular demise. Finally, we strongly recommend to exclude the presence of apoptotic death indicators, and most importantly to differentiate the observed phenotype from secondary necrosis.
–
As in higher eukaryotic cells, in yeast, ACD may be triggered upon the challenge to extremely detrimental conditions. Thus, agents like hydrogen peroxide, acetic acid, amphotericin B, or several metals that are pro-apoptotic at low doses may induce necrosis at high concentrations [125][199][223][224]. We assume that necrosis is the consequence of radical cellular damage in most of these cases, and hence a bona fide instance of ACD. This is in line with the concept that not only the type but also the intensity of a given perturbation determines the form of death [91][225].
–
As mentioned above, yeast can undergo regulated necrosis, reminiscent of the RCD instances detected in human cells [226]. Indeed, genetic and chemical manipulations demonstrate that yeast necrosis can be inhibited, at least in some settings, indicating that it results from the activation of a molecular mechanism. In order to differentiate regulated from accidental necrosis, it is necessary to test whether a pharmacological or genetic intervention is capable of inhibiting necrosis in the scenario that is being studied. Known necrosis-modulatory approaches include the exogenous administration of the naturally occurring polyamine spermidine, which can specifically reduce primary necrotic cell death in the context of chronological aging [160]. A similar outcome can be obtained by genetic modulation of polyamine biosynthesis [160]. In addition, the proteolytically inactive propeptide of the vacuolar endoprotease Pep4, the homolog of human cathepsin D, has been shown to mediate antinecrotic effects. Accordingly, prolonged overexpression of Pep4 (or its propeptide) can extend chronological lifespan via specific inhibition of necrosis [104][227]. Intriguingly, the antinecrotic function of Pep4 depends on polyamine biosynthesis [104]. In fact, further vacuolar factors as well as other organelles, e.g., peroxisomes, might be connected to regulated necrosis, but this requires further investigation [228][229][230][231][232].
–
Under certain circumstances, regulated necrosis in mammalian cells may be mechanistically linked to primary Δψm dissipation [233][234], and such a mitochondrial permeability transition (MPT)-driven necrosis is connected to a series of pathological conditions [235]. In yeast, necrotic cell death also seems to depend on mitochondria in several settings [149][220][202]. In addition, recent reports show that necrotic cell death upon a lipotoxic insult requires a functional Rim101 signaling cascade that involves the calpain-like protease Rim13/Cpl1 for lethal execution [236][237]. To interrogate a possible case of regulated necrosis, it is thus advisable to evaluate a possible mitochondrial involvement. For that purpose, it would be indicated to examine whether necrosis is diminished upon abrogation of mitochondrial function, e.g., in a ρ0 strain (which lacks mitochondrial DNA). However, as previously mentioned, mitochondria are the main executors of apoptotic cell death. Thus, mitochondrial dependence cannot be used as a sole determinant to characterize regulated necrosis and must be accompanied by a set of other assays that demonstrate the primary necrotic nature of cell death. Of note, several known mammalian mediators of regulated necrosis have homologs in yeast, including cathepsins, cyclophilin D, calpains, Hsp90, or protein kinase A, among others [226], but only a few of them have been examined in this context [103][238]. It will be interesting to see whether these factors possess a conserved necrotic function in yeast, which would expand the possibilities to determine bona fide regulated necrosis. Similarly, it remains to be seen whether known inhibitors of regulated necrosis in mammals also interfere with some cell death scenarios in yeast as well [239].
–
A number of questions remain to be answered with regard to the actual existence of a necrotic RCD subroutine in yeast. In mammals, regulated necrosis plays a number of key roles, most prominently due to its immunogenic nature, for instance upon pathogen infection [226]. Such interaction with the immune system, however, is a feature of complex multicellular organisms. Nonetheless, several physiological scenarios in which regulated necrosis seems to be instrumental for yeast, provide a teleological explanation for its existence in a unicellular organism. During chronological aging, for instance, yeast cells die exhibiting markers of both early/late apoptosis and primary necrosis [61][75][160]. Interestingly, the fraction of cells dying by primary necrosis actually represents the majority of the dying population that is reduced upon a cytoprotective intervention, at least via polyamine-mediated lifespan extension [104][160]. Another example is the necrotic death of the meiotic mother cell during the terminal stages of gametogenesis (sporulation) [149]. In this setting, necrosis occurs after the spores have reached the final phases of development, suggesting a controlled coordination that allows for gamete differentiation prior to the elimination of the mother cell. This might well constitute an instance of necrotic PCD, reinforcing the notion that yeast populations must be seen as a multicellular community of genetically identical cells that responds to selective pressures by ensuring the long-term survival of at least one clonal individual. Therefore, it is conceivable that regulated necrosis might participate in cell-to-cell communication via the inevitable release of intracellular contents, as this is the case in higher eukaryotes [226]. Such hypothetical necrosis-related quorum-sensing molecules, however, are yet to be identified in yeast.
–
In human cells, different types of regulated necrosis have been defined, with MPT-driven regulated necrosis and necroptosis among the most extensively studied forms [240][241]. In yeast, mechanistic insights into the control of necrosis are still very limited at this point. Thus, we strongly discourage the use of neologisms to avoid confusion. Instead, we propose to employ the term “regulated necrosis” to describe any genetically controlled form of necrosis in yeast (or “programmed necrosis” if it is a form of PCD). Further research into the molecular activators, transducers and executioners of regulated necrosis in yeast will reveal whether potentially different subroutines of the process exist.
OTHER RCD TYPES
In mammalian cells, a series of other RCD modalities have been defined. Macroautophagy (hereafter referred to as autophagy) is a conserved catabolic process that orchestrates the digestion of intracellular material (e.g., protein aggregates, organelles) in the vacuole. During autophagy, double-membraned vesicles (so-called autophagosomes) form and engulf cytoplasmic components, followed by the fusion of autophagosomes with the vacuole, where the cargo is degraded and the resulting macromolecules are released into the cytoplasm for reuse [16][242]. Thus, autophagy is predominantly a cell survival mechanism (see below). Historically, though, “autophagic cell death” (ATCD) was one of the three distinct cell death manifestations (besides apoptosis and necrosis) that were described for human cells based on morphological criteria [243]. Although this original description did not indicate any functional connection, it became a widespread belief that ATCD would point to cell death as a mechanistic outcome of autophagy. The term ATCD has indeed been extensively misused to describe cell death instances that occur in the presence of autophagic markers, instead of testing an actual dependency on the process and/or its molecular machinery, i.e., assessing the retardation of cell death via pharmacological or genetic inhibition of autophagy [242]. In fact, the NCCD has recently agreed to identify such forms of cell death as “autophagy-dependent cell death” (ADCD) [95]. ADCD can in principle describe (i) cell death dependent on the autophagic machinery (in its whole, or parts thereof) and (ii) cell death dependent on actual autophagic degradation. Indeed, (i) components of the autophagic machinery have been etiologically implicated in specific settings of RCD in Drosophila melanogaster and human cells [225][244][245][246][247][248]. In these contexts, the molecular apparatus for autophagy contributes to cellular demise. To our knowledge, however, there is no study in which cell death has been directly linked to (ii) a functional autophagic flux. Thus, we surmise that most cases of ADCD rather depend on components of the autophagic machinery than on autophagic responses. In fact, the molecular machinery of ADCD and adaptive autophagy partially differ (at least in D. melanogaster) [249][250].
–
In yeast, the term ATCD has been used to describe cellular demise occurring under specific external stress conditions like zinc-induced cell death [251], heterologous expression of human α-synuclein [252] or human p53 [253] as well as internal deficiencies like defects in inorganic pyrophosphatases [254]. Following the recent proposition by the NCCD, we favor the use of the term ADCD (instead of ATCD) in yeast, as well. Again, ADCD should be used to describe cell death only when autophagy (or at least two proteins from the autophagic machinery, see below) has been experimentally given an etiological implication in the process. As a note of caution, it is important to underscore that the term ADCD should be avoided if the autophagic machinery (or components thereof) is activated parallel to (rather than triggering) RCD or if it promotes other RCD subroutines [95]. In fact, in most known cases from yeast to human, autophagy acts as a cytoprotective response to detrimental stress conditions, in which it disposes damaged cellular material [37][255][256]. Accordingly, cell death is rather accelerated than repressed upon inhibition of autophagy in both human cells and yeast [257][258][259]. In fact, and despite the evidence for autophagy activation in the course of cell death (see above), the very existence of ADCD as an actual cell death type has been questioned [260][261]. In any case, cell death may often be preceded or accompanied by autophagy markers, probably mirroring the final effort of dying cells to counteract a lethal stress. Thus, in most cases, cells showing biomarkers of autophagy might be dying with, and not by, autophagy. We thus consider that the use of the term ADCD should be used with utmost care, taking into account the aforementioned NCCD recommendations [95].
–
A number of microscopic, biochemical and enzymatic assays are available and established [262][263][264] to determine autophagic flux, i.e., the progression through the pathway and thus its degradation activity [262][265]. One of the most common methods to measure autophagic flux in yeast is to evaluate the vacuolar processing (or GFP liberation) of N-terminally GFP-tagged Atg8, a central modulator of autophagosome formation, and its delivery to the vacuole, via fluorescence or immunoblot analysis [266][267][268][269][270]. Other widely used assays include assessing the autophagy-dependent activity of a modified version of the vacuolar alkaline phosphatase Pho8 via a specific enzymatic assay [118][271] or monitoring the pH-change of cellular compartments upon delivery of pH-sensitive fluorescent proteins to the vacuole (such as Rosella) [272]. However, such quantitative assessments – while necessary – are not sufficient to characterize ADCD: for that purpose, a functional dependency on the autophagic machinery (or components thereof) must be concluded, as mentioned above. Thus, all cases of cell death that are accompanied by autophagic markers, but cannot be suppressed or retarded by inhibiting the (at least parts of) the molecular apparatus of autophagy should not be considered as ADCD.
–
The causative implication of autophagy in cell death may be explored by deletion of autophagy-related (ATG) genes, which are the key orchestrators of the process [262]. However, ATGs may have autophagy-unrelated functions as well [273]. Thus, akin to the recommendations for higher eukaryotes [242], we suggest testing at least two (and better more) distinct ATG deletions to assess dependency on the autophagic machinery. Inhibitory components of the autophagic apparatus can also be targeted, e.g., by constitutively activating the TOR complex 1 or the RAS/PKA signaling pathway, resulting in autophagy suppression [274][275][276][277]. As mentioned above, dependence of cell death on the molecular machinery of autophagy (in its whole, or parts thereof) does not imply cell death to be dependent on autophagic degradation. To evaluate if the autophagic response is implicated in the lethal execution, one may take advantage of chemical inhibition [262][278]. Vacuolar proteolysis can be blocked through direct inhibition of proteases either genetically (e.g., by deleting PEP4 or PRB1) or pharmacologically (e.g., by addition of pepstatin A, E-64D, leupeptin alone or in combination) as well as by neutralizing the vacuolar pH (e.g., by means of chloroquine) [262][265]. In yeast, chemical inhibition of autophagosome formation (as it is commonly applied in mammals using specific suppressors of phosphatidylinositol 3-kinase) is not typically employed, since substantially higher concentrations of these drugs are often needed [262][265]. In fact, genetic approaches are generally favored in the ADCD field due to insufficient specificity of most pharmacological autophagy inhibitors [242].
–
The expression “mitotic catastrophe” (MC) was first employed to illustrate the lethal phenotype of a temperature-sensitive fission yeast mutant strain that enters mitosis prematurely without effectively completing it [11]. The term MC has since been most frequently used to define cell death that occurs upon aberrant mitosis [94], which is frequently accompanied by gross nuclear alterations. In yeast, as in mammals, it may result from genome instability, microtubule destabilization, DNA damage, or alterations in cell cycle checkpoints [279][280][281][282]. Intriguingly, yeast RCD has been connected to most of these features [63][283][284][285]. It will be interesting to follow whether known MC scenarios culminate in specific RCD subroutines.
–
In mammalian cells, death following mitotic aberrations can, indeed, be either apoptotic or necrotic [286]. Since mitotic defects may contribute to malignant transformation in the mammalian system, MC can be viewed as an oncosuppressive mechanism that operates via cell death or senescence [94][286]. In fact, suppression of MC provokes tumorigenesis and cancer progression in mammals [287]. By analogy, MC in yeast might be a mechanism to eliminate mitosis-incompetent and thus unfit cells from the population. Adhering to the recommendations by the NCCD [94], we thus propose to use the term MC as an independent molecular avenue that precedes RCD, but does not constitute a bona fide cell death executioner mechanism by itself [287].
–
A series of other cell death subroutines have been defined in human cells that, however, are restricted to specific cell types and thus do not apply to yeast.
RCD IN OTHER YEASTS AND FILAMENTOUS FUNGI
As previously mentioned, yeast cell death has been most extensively studied in S. cerevisiae. However, other yeast species have been shown to share similar cell death characteristics and also bear a set of comparable cell death subroutines. Thus, we propose to extend the above-described recommendations formulated above to all yeast species.
–
Schizosaccharomyces pombe (fission yeast) has been shown to express an RCD machinery that responds to various stimuli. These include physiological triggers such as aging, defects like the abnormal metabolism of intracellular lipids [288][289][290][291], and a number of insults, including ER stress [292], inositol starvation [289][290] or the heterologous expression of several metazoan apoptotic effectors, e.g., BAX and BAK [293]. All of these stimuli converge on the activation of apoptosis. Of note, according to our definitions, neither regulated necrosis nor ADCD have been demonstrated in S. pombe (yet). Among the described S. pombe apoptosis executors are the chaperone Cnx1 (calnexin) and the metacaspase Pca1 [292][294]. Pca1 is involved in the apoptotic response to inositol starvation [292][294] and lipid-induced, non-apoptotic cell death in minimal medium. Conversely, Pca1 does not seem to play any role during apoptosis induced by ER stress [292], valproic acid treatment [295], or lipotoxic stress in minimal medium [289]. S. pombe apoptosis is expected to involve additional players, as there is evidence for the presence of different factors in fission yeast that are homologous to effectors of S. cerevisiae apoptosis, including the protease Nma111 [296], Aif1 [297] or endonuclease G [298].
–
The major opportunistic human pathogen Candida albicans, which has become a molecular genetics model to study pathogenicity, virulence and fungal development [299][300], can also undergo apoptosis following the exposure to a plethora of different agents [224][301][302][303]. To date, no RCD subroutines other than apoptosis have been described. Interestingly, apoptosis can occur in both the blastospore and the hyphal form of this organism [302] as well as in Candida biofilms, which are highly tolerant to standard antimycotics and hence difficult to eradicate. Exploiting the apoptosis machinery in cells constituting biofilms may pave the way to their effective eradication, and hence limit the incidence of indwelling device-associated infections (IDAIs) [304][305][306]. C. albicans also harbors a gene encoding a metacaspase (CaMCA1) [307], which mediates apoptosis, for instance, upon treatment with farnesol [308], caspofungin [309], and micafungin [213] or upon interaction with murine macrophages [308][310]. Conversely, CaMca1 is not involved in other apoptotic settings like exposure to the plant defensin RsAFP2 [311]. The Ras–cAMP–PKA signaling pathway [312] and the bZip transcription factor Cap1 [313][314] have also been implicated in distinct apoptotic scenarios. Finally, other closely related Candida species, e.g., Candida glabrata [315], Candida krusei [316], Candida dubliniensis [317], Candida tropicalis [318], or Candida parapsilosis [213][319], have been reported to exhibit apoptotic markers upon lethal challenge.
–
Cryptococcus neoformans, an important pathogen of immunocompromised and immunocompetent patients, also undergoes apoptosis [320][321], with apoptosis-inducing factor and two metacaspases independently orchestrating this lethal subroutine [321]. At least one other Cryptococcus genus member, Cryptococcus laurentii, has also been shown to respond to some stimuli with apoptotic RCD [322]. Furthermore, a number of other yeast species, e.g., Kluyveromyces lactis [323][324], Pichia pastoris [325], Rhodotorula glutinis [326], or Zygosaccharomyces bailii [327][328], may develop signs of apoptosis under certain conditions. We surmise that similar lethal programs are to be discovered in other yeast species. In fact, such discoveries and further characterization of both identified and yet uncovered RCD programs are expected to follow in the near future, given that antifungal therapeutics for medical and industrial purposes may increasingly rely on targeting the yeast RCD machinery [329][330]. We thus suggest adopting the recommendations formulated above for the description of cell death in all types of yeasts.
–
It should be noted that a growing body of work is addressing RCD in multicellular fungi. A major human pathogenic fungus that causes life-threatening disease is Aspergillus fumigatus, which has also been demonstrated to undergo apoptosis under certain conditions [331][332]. The genome of A. fumigatus codes for two metacaspases (CasA and CasB), whose relative contribution to cell death seems to depend on the scenario [332][333][334]. In fact, other fungal proteases might also exert metacaspase activities that are relevant for cell viability and/or survival [334]. Aspergillus nidulans is another member of the Aspergillus spp. that has been demonstrated to undergo RCD [335][336]. The genome of A. nidulans appears to code for an apoptotic machinery with relevant players like apoptosis-inducing factor and two putative metacaspases [336][337]. Another filamentous fungus, Podospora anserina, is used as an aging model that incorporates crucial apoptotic factors, including two metacaspases (PaMCA1 and PaMCA2) and at least five Aif members, of which only mitochondrial (but not cytosolic) isoforms seem to be relevant for aging-driven RCD [338][339]. A role for the P. anserina cyclophilin D ortholog in RCD [340][341][342] as well as for autophagy in aging and lifespan control of P. anserina [343][344] have been reported. Further instances of fungal RCD [303][345] have been documented in Paracoccidioides brasiliensis [346], Colletotrichum gloeosporioides [347], Fusarium oxysporum [348], Fusarium graminearum [349], Mucor racemosus [350], Botrytis cinerea [351], Penicillium expansum [352], Rhizopus oryzae [353], Scedosporium prolificans [354] and Neurospora crassa [355]. As multicellular organisms, filamentous fungi have developed programs that are reminiscent of organismal RCD. For instance, several putative homologs of factors relevant for animal apoptotic control that are not found in unicellular yeast are present in the genomes of filamentous species [356]. Thus, multicellular fungi may have complex traits not present in yeasts that may add to the criteria and definitions presented herein.
CONCLUDING REMARKS
The impact of yeast (and other fungi, including filamentous species) on our lives at multiple socioeconomic, scientific and medical levels emphasizes the importance of decoding the mechanisms that determine its survival and control its demise. Therefore, the molecular comprehension and potential manipulation of yeast cell death hold major promise for biotechnological and biomedical applications. We anticipate that numerous fields might benefit from the possibility to modulate yeast cell death. For instance, the productivity of yeast during large-scale processes in the pharmaceutical and industrial arenas largely depends on its viability and ultimately on its tolerance to stress and its demise in stationary cultures. Also, novel pharmacological approaches that specifically target the RCD machinery of yeast pathogens may bypass the ever-increasing resistance to classical antimycotics, which is an emerging public health problem. Other medical manipulations of yeast RCD are also conceivable, e.g., strategies to intervene on pathogenic deviations of the mycobiome. Finally, yeast will continue to help the community in deciphering eukaryotic cell death pathways as it serves as an important model for human disease. Given its power to study the relationship between genotype and phenotype, we expect to gain further insights from yeast to identify actionable targets that may be subjected to pharmacological (drug discovery) or genetical manipulation.
–
For all these reasons, it is now imperative to set the standards for defining and studying cell death in yeast. That said, we want to emphasize that the present set of recommendations should be taken – as any scientific overview – as a snapshot of the current knowledge, rather than as a definitive compilation. Indeed, as research continues, we surmise that the present guidelines will have to be extended and revised. For instance, other nuanced changes to – or even novel types of – RCD may emerge from continued efforts to characterize the multicellular character of yeast populations, including but not restricted to uncovering intercellular communication, interaction between populations or cellular differentiation within colonies and biofilms. Still, neologisms should be introduced with care and only when the characterization of a lethal process that bears new functional and biochemical aspects requires it. Otherwise, new expressions should be avoided to limit confusion.
–
Another crucial point is to acknowledge the inherent complexity and dynamic nature of RCD in general and its different subroutines in particular. In fact, it is the crosstalk between pro-life and pro-death signals that determines cellular fate, and the activation of pro-survival pathways (such as autophagy) may often accompany lethal signals. Also, stress conditions may activate different RCD subroutines that can be interconnected or may occur independently, sequentially, or in parallel. Indeed, the inhibition of one specific RCD modality might trigger backup mechanisms that still ensure cell death execution. It is thus important to keep these points in mind when classifying a lethal phenotype.
–
Altogether, the present guidelines attempt to unify the nomenclature and definition of yeast cell death modalities and – in our opinion – will help other fields of unicellular research (e.g., bacteriology, parasitology, etc.) to establish their set of recommendations using the present one as a basis. We are convinced that some degree of linguistic and experimental standardization is necessary for facilitating communication among researchers, especially at a point where the existence of yeast RCD is scientifically accepted and its socioeconomical impact is ever growing.
References
- I. Masneuf-Pomarede, M. Bely, P. Marullo, and W. Albertin, "The Genetics of Non-conventional Wine Yeasts: Current Knowledge and Future Challenges", Frontiers in Microbiology, vol. 6, 2016. http://dx.doi.org/10.3389/fmicb.2015.01563
- A.S. Zaky, G.A. Tucker, Z.Y. Daw, and C. Du, "Marine yeast isolation and industrial application", FEMS Yeast Research, vol. 14, pp. 813-825, 2014. http://dx.doi.org/10.1111/1567-1364.12158
- . BCC Research LLC, "Yeasts, yeast extracts, autolysates and related products: the global market (CHM053C)", 2014.
- M.J. Suhr, and H.E. Hallen-Adams, "The human gut mycobiome: pitfalls and potentials--a mycologists perspective", Mycologia, vol. 107, pp. 1057-1073, 2015. http://dx.doi.org/10.3852/15-147
- J.J. Limon, J.H. Skalski, and D.M. Underhill, "Commensal Fungi in Health and Disease", Cell Host & Microbe, vol. 22, pp. 156-165, 2017. http://dx.doi.org/10.1016/j.chom.2017.07.002
- E.J. Polvi, X. Li, T.R. O’Meara, M.D. Leach, and L.E. Cowen, "Opportunistic yeast pathogens: reservoirs, virulence mechanisms, and therapeutic strategies", Cellular and Molecular Life Sciences, vol. 72, pp. 2261-2287, 2015. http://dx.doi.org/10.1007/s00018-015-1860-z
- G.D. Brown, D.W. Denning, N.A.R. Gow, S.M. Levitz, M.G. Netea, and T.C. White, "Hidden Killers: Human Fungal Infections", Science Translational Medicine, vol. 4, 2012. http://dx.doi.org/10.1126/scitranslmed.3004404
- M.C. Fisher, D.A. Henk, C.J. Briggs, J.S. Brownstein, L.C. Madoff, S.L. McCraw, and S.J. Gurr, "Emerging fungal threats to animal, plant and ecosystem health", Nature, vol. 484, pp. 186-194, 2012. http://dx.doi.org/10.1038/nature10947
- D. Botstein, and G.R. Fink, "Yeast: An Experimental Organism for 21st Century Biology", Genetics, vol. 189, pp. 695-704, 2011. http://dx.doi.org/10.1534/genetics.111.130765
- T. Evans, E.T. Rosenthal, J. Youngblom, D. Distel, and T. Hunt, "Cyclin: a protein specified by maternal mRNA in sea urchin eggs that is destroyed at each cleavage division.", Cell, 1983. http://www.ncbi.nlm.nih.gov/pubmed/6134587
- P. Russell, and P. Nurse, "cdc25+ functions as an inducer in the mitotic control of fission yeast.", Cell, 1986. http://www.ncbi.nlm.nih.gov/pubmed/3955656
- L.H. Hartwell, and T.A. Weinert, "Checkpoints: controls that ensure the order of cell cycle events.", Science (New York, N.Y.), 1989. http://www.ncbi.nlm.nih.gov/pubmed/2683079
- K. Nasmyth, "At the heart of the budding yeast cell cycle", Trends in Genetics, vol. 12, pp. 405-412, 1996. http://dx.doi.org/10.1016/0168-9525(96)10041-X
- M. Polymenis, . , R. Aramayo, and . , "Translate to divide: сontrol of the cell cycle by protein synthesis", Microbial Cell, vol. 2, pp. 94-104, 2015. http://dx.doi.org/10.15698/mic2015.04.198
- J. Jimenez, . , S. Bru, M. Ribeiro, and J. Clotet, "Live fast, die soon: cell cycle progression and lifespan in yeast cells", Microbial Cell, vol. 2, pp. 62-67, 2015. http://dx.doi.org/10.15698/mic2015.03.191
- Z. Yin, C. Pascual, and D. Klionsky, "Autophagy: machinery and regulation", Microbial Cell, vol. 3, pp. 588-596, 2016. http://dx.doi.org/10.15698/mic2016.12.546
- A. Zimmermann, K. Kainz, A. Andryushkova, S. Hofer, F. Madeo, and D. Carmona-Gutierrez, "Autophagy: one more Nobel Prize for yeast", Microbial Cell, vol. 3, pp. 579-581, 2016. http://dx.doi.org/10.15698/mic2016.12.544
- M. Tsukada, and Y. Ohsumi, "Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae.", FEBS letters, 1993. http://www.ncbi.nlm.nih.gov/pubmed/8224160
- K. Takeshige, M. Baba, S. Tsuboi, T. Noda, and Y. Ohsumi, "Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction.", The Journal of cell biology, 1992. http://www.ncbi.nlm.nih.gov/pubmed/1400575
- P. Spincemaille, . , B. Cammue, K. Thevissen, and . , "Sphingolipids and mitochondrial function, lessons learned from yeast", Microbial Cell, vol. 1, pp. 210-224, 2014. http://dx.doi.org/10.15698/mic2014.07.156
- L. Galluzzi, O. Kepp, and G. Kroemer, "Mitochondrial regulation of cell death: a phylogenetically conserved control", Microbial Cell, vol. 3, pp. 101-108, 2016. http://dx.doi.org/10.15698/mic2016.03.483
- M. Opalinska, . , C. Meisinger, and . , "Mitochondrial protein import under kinase surveillance", Microbial Cell, vol. 1, pp. 51-57, 2014. http://dx.doi.org/10.15698/mic2014.01.127
- W. Neupert, and J.M. Herrmann, "Translocation of Proteins into Mitochondria", Annual Review of Biochemistry, vol. 76, pp. 723-749, 2007. http://dx.doi.org/10.1146/annurev.biochem.76.052705.163409
- V. Peleh, . , J. Riemer, A. Dancis, J. Herrmann, . , and . , "Protein oxidation in the intermembrane space of mitochondria is substrate-specific rather than general", Microbial Cell, vol. 1, pp. 81-93, 2014. http://dx.doi.org/10.15698/mic2014.01.130
- A. Chacinska, C.M. Koehler, D. Milenkovic, T. Lithgow, and N. Pfanner, "Importing Mitochondrial Proteins: Machineries and Mechanisms", Cell, vol. 138, pp. 628-644, 2009. http://dx.doi.org/10.1016/j.cell.2009.08.005
- A. Bachmair, D. Finley, and A. Varshavsky, "In Vivo Half-Life of a Protein Is a Function of Its Amino-Terminal Residue", Science, vol. 234, pp. 179-186, 1986. http://dx.doi.org/10.1126/science.3018930
- P. Novick, C. Field, and R. Schekman, "Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway.", Cell, 1980. http://www.ncbi.nlm.nih.gov/pubmed/6996832
- I.C. Griff, R. Schekman, J.E. Rothman, and C.A. Kaiser, "The yeast SEC17 gene product is functionally equivalent to mammalian alpha-SNAP protein.", The Journal of biological chemistry, 1992. http://www.ncbi.nlm.nih.gov/pubmed/1601878
- A. Skoneczna, A. Kaniak, and M. Skoneczny, "Genetic instability in budding and fission yeast—sources and mechanisms", FEMS Microbiology Reviews, vol. 39, pp. 917-967, 2015. http://dx.doi.org/10.1093/femsre/fuv028
- V. Tosato, and C. Bruschi, "Per aspera ad astra: When harmful chromosomal translocations become a plus value in genetic evolution. Lessons from Saccharomyces cerevisiae.", Microbial Cell, vol. 2, pp. 363-375, 2015. http://dx.doi.org/10.15698/mic2015.10.230
- B. Fahrenkrog, "Histone modifications as regulators of life and death in Saccharomyces cerevisiae", Microbial Cell, vol. 3, pp. 1-13, 2016. http://dx.doi.org/10.15698/mic2016.01.472
- G. Janssens, and L. Veenhoff, "Evidence for the hallmarks of human aging in replicatively aging yeast", Microbial Cell, vol. 3, pp. 263-274, 2016. http://dx.doi.org/10.15698/mic2016.07.510
- A.X. Santos, and H. Riezman, "Yeast as a model system for studying lipid homeostasis and function", FEBS Letters, vol. 586, pp. 2858-2867, 2012. http://dx.doi.org/10.1016/j.febslet.2012.07.033
- D. Petranovic, K. Tyo, G.N. Vemuri, and J. Nielsen, "Prospects of yeast systems biology for human health: integrating lipid, protein and energy metabolism", FEMS Yeast Research, vol. 10, pp. 1046-1059, 2010. http://dx.doi.org/10.1111/j.1567-1364.2010.00689.x
- S. Schroeder, . , A. Zimmermann, D. Carmona-Gutierrez, T. Eisenberg, C. Ruckenstuhl, A. Andryushkova, T. Pendl, A. Harger, F. Madeo, and . , "Metabolites in aging and autophagy", Microbial Cell, vol. 1, pp. 110-114, 2014. http://dx.doi.org/10.15698/mic2014.04.142
- M. Conrad, J. Schothorst, H.N. Kankipati, G. Van Zeebroeck, M. Rubio-Texeira, and J.M. Thevelein, "Nutrient sensing and signaling in the yeastSaccharomyces cerevisiae", FEMS Microbiology Reviews, vol. 38, pp. 254-299, 2014. http://dx.doi.org/10.1111/1574-6976.12065
- R. de Cabo, D. Carmona-Gutierrez, M. Bernier, M. Hall, and F. Madeo, "The Search for Antiaging Interventions: From Elixirs to Fasting Regimens", Cell, vol. 157, pp. 1515-1526, 2014. http://dx.doi.org/10.1016/j.cell.2014.05.031
- L. Fontana, L. Partridge, and V.D. Longo, "Extending Healthy Life Span—From Yeast to Humans", Science, vol. 328, pp. 321-326, 2010. http://dx.doi.org/10.1126/science.1172539
- A. Shrestha, and L. Megeney, "Yeast proteinopathy models: a robust tool for deciphering the basis of neurodegeneration", Microbial Cell, vol. 2, pp. 458-465, 2015. http://dx.doi.org/10.15698/mic2015.12.243
- S. Tenreiro, M.C. Munder, S. Alberti, and T.F. Outeiro, "Harnessing the power of yeast to unravel the molecular basis of neurodegeneration", Journal of Neurochemistry, vol. 127, pp. 438-452, 2013. http://dx.doi.org/10.1111/jnc.12271
- R.J. Braun, S. Büttner, J. Ring, G. Kroemer, and F. Madeo, "Nervous yeast: modeling neurotoxic cell death", Trends in Biochemical Sciences, vol. 35, pp. 135-144, 2010. http://dx.doi.org/10.1016/j.tibs.2009.10.005
- A. Ruetenik, A. Ocampo, K. Ruan, Y. Zhu, C. Li, G. Zhai, and A. Barrientos, "Attenuation of polyglutamine-induced toxicity by enhancement of mitochondrial OXPHOS in yeast and fly models of aging", Microbial Cell, vol. 3, pp. 338-351, 2016. http://dx.doi.org/10.15698/mic2016.08.518
- R.B. Wickner, "[URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae.", Science (New York, N.Y.), 1994. http://www.ncbi.nlm.nih.gov/pubmed/7909170
- S. Speldewinde, and C. Grant, "The frequency of yeast [PSI+] prion formation is increased during chronological ageing", Microbial Cell, vol. 4, pp. 127-132, 2017. http://dx.doi.org/10.15698/mic2017.04.568
- S. Park, K. Ratia, M. Ba, M. Valencik, and S. Liebman, "Inhibition of Aβ42 oligomerization in yeast by a PICALM ortholog and certain FDA approved drugs", Microbial Cell, vol. 3, pp. 53-64, 2016. http://dx.doi.org/10.15698/mic2016.02.476
- J.J. Heinisch, and R. Brandt, "Signaling pathways and posttranslational modifications of tau in Alzheimer's disease: the humanization of yeast cells", Microbial Cell, vol. 3, pp. 135-146, 2016. http://dx.doi.org/10.15698/mic2016.04.489
- S. Alves, L. Castro, M.S. Fernandes, R. Francisco, P. Castro, M. Priault, S.R. Chaves, M.P. Moyer, C. Oliveira, R. Seruca, M. Côrte-Real, M.J. Sousa, and A. Preto, "Colorectal cancer-related mutant KRAS alleles function as positive regulators of autophagy", Oncotarget, vol. 6, pp. 30787-30802, 2015. http://dx.doi.org/10.18632/oncotarget.5021
- G. Li, M. Poulsen, C. Fenyvuesvolgyi, Y. Yashiroda, M. Yoshida, J.M. Simard, R.C. Gallo, and R.Y. Zhao, "Characterization of cytopathic factors through genome-wide analysis of the Zika viral proteins in fission yeast", Proceedings of the National Academy of Sciences, vol. 114, 2017. http://dx.doi.org/10.1073/pnas.1619735114
- R.Y. Zhao, "Yeast for virus research", Microbial Cell, vol. 4, pp. 311-330, 2017. http://dx.doi.org/10.15698/mic2017.10.592
- N. Guaragnella, V. Palermo, A. Galli, L. Moro, C. Mazzoni, and S. Giannattasio, "The expanding role of yeast in cancer research and diagnosis: insights into the function of the oncosuppressors p53 and BRCA1/2", FEMS Yeast Research, vol. 14, pp. 2-16, 2013. http://dx.doi.org/10.1111/1567-1364.12094
- R. Diaz-Ruiz, M. Rigoulet, and A. Devin, "The Warburg and Crabtree effects: On the origin of cancer cell energy metabolism and of yeast glucose repression", Biochimica et Biophysica Acta (BBA) - Bioenergetics, vol. 1807, pp. 568-576, 2011. http://dx.doi.org/10.1016/j.bbabio.2010.08.010
- S. Tenreiro, V. Franssens, J. Winderickx, and T.F. Outeiro, "Yeast models of Parkinson’s disease-associated molecular pathologies", Current Opinion in Genetics & Development, vol. 44, pp. 74-83, 2017. http://dx.doi.org/10.1016/j.gde.2017.01.013
- C. Ruckenstuhl, S. Büttner, D. Carmona-Gutierrez, T. Eisenberg, G. Kroemer, S.J. Sigrist, K. Fröhlich, and F. Madeo, "The Warburg Effect Suppresses Oxidative Stress Induced Apoptosis in a Yeast Model for Cancer", PLoS ONE, vol. 4, pp. e4592, 2009. http://dx.doi.org/10.1371/journal.pone.0004592
- K. Peeters, F. Van Leemputte, B. Fischer, B.M. Bonini, H. Quezada, M. Tsytlonok, D. Haesen, W. Vanthienen, N. Bernardes, C.B. Gonzalez-Blas, V. Janssens, P. Tompa, W. Versées, and J.M. Thevelein, "Fructose-1,6-bisphosphate couples glycolytic flux to activation of Ras", Nature Communications, vol. 8, 2017. http://dx.doi.org/10.1038/s41467-017-01019-z
- T. Rajakumar, A.B. Munkacsi, and S.L. Sturley, "Exacerbating and reversing lysosomal storage diseases: from yeast to humans", Microbial Cell, vol. 4, pp. 278-293, 2017. http://dx.doi.org/10.15698/mic2017.09.588
- S. Stekovic, C. Ruckenstuhl, P. Royer, C. Winkler-Hermaden, D. Carmona-Gutierrez, K. Fröhlich, G. Kroemer, and F. Madeo, "The neuroprotective steroid progesterone promotes mitochondrial uncoupling, reduces cytosolic calcium and augments stress resistance in yeast cells", Microbial Cell, vol. 4, pp. 191-199, 2017. http://dx.doi.org/10.15698/mic2017.06.577
- S. Bach, N. Talarek, T. Andrieu, J. Vierfond, Y. Mettey, H. Galons, D. Dormont, L. Meijer, C. Cullin, and M. Blondel, "Isolation of drugs active against mammalian prions using a yeast-based screening assay", Nature Biotechnology, vol. 21, pp. 1075-1081, 2003. http://dx.doi.org/10.1038/nbt855
- M.J. Lista, C. Voisset, M. Contesse, G. Friocourt, C. Daskalogianni, F. Bihel, R. Fåhraeus, and M. Blondel, "The long‐lasting love affair between the budding yeast Saccharomyces cerevisiae and the Epstein‐Barr virus", Biotechnology Journal, vol. 10, pp. 1670-1681, 2015. http://dx.doi.org/10.1002/biot.201500161
- M.J. Lista, R.P. Martins, G. Angrand, A. Quillévéré, C. Daskalogianni, C. Voisset, M. Teulade-Fichou, R. Fåhraeus, and M. Blondel, "A yeast model for the mechanism of the Epstein‐Barr virus immune evasion identifies a new therapeutic target to interfere with the virus stealthiness", Microbial Cell, vol. 4, pp. 305-307, 2017. http://dx.doi.org/10.15698/mic2017.09.590
- M.J. Lista, R.P. Martins, O. Billant, M. Contesse, S. Findakly, P. Pochard, C. Daskalogianni, C. Beauvineau, C. Guetta, C. Jamin, M. Teulade-Fichou, R. Fåhraeus, C. Voisset, and M. Blondel, "Nucleolin directly mediates Epstein-Barr virus immune evasion through binding to G-quadruplexes of EBNA1 mRNA", Nature Communications, vol. 8, 2017. http://dx.doi.org/10.1038/ncomms16043
- D. Carmona‐Gutierrez, and S. Büttner, "The many ways to age for a single yeast cell", Yeast, vol. 31, pp. 289-298, 2014. http://dx.doi.org/10.1002/yea.3020
- D. Carmona-Gutierrez, T. Eisenberg, S. Büttner, C. Meisinger, G. Kroemer, and F. Madeo, "Apoptosis in yeast: triggers, pathways, subroutines", Cell Death & Differentiation, vol. 17, pp. 763-773, 2010. http://dx.doi.org/10.1038/cdd.2009.219
- F. Madeo, E. Fröhlich, and K.U. Fröhlich, "A yeast mutant showing diagnostic markers of early and late apoptosis.", The Journal of cell biology, 1997. http://www.ncbi.nlm.nih.gov/pubmed/9348289
- F. Madeo, D. Carmona-Gutierrez, J. Ring, S. Büttner, T. Eisenberg, and G. Kroemer, "Caspase-dependent and caspase-independent cell death pathways in yeast", Biochemical and Biophysical Research Communications, vol. 382, pp. 227-231, 2009. http://dx.doi.org/10.1016/j.bbrc.2009.02.117
- T. Eisenberg, S. Büttner, G. Kroemer, and F. Madeo, "The mitochondrial pathway in yeast apoptosis", Apoptosis, vol. 12, pp. 1011-1023, 2007. http://dx.doi.org/10.1007/s10495-007-0758-0
- P. Polcic, . , P. Jaka, and M. Mentel, "Yeast as a tool for studying proteins of the Bcl-2 family", Microbial Cell, vol. 2, pp. 74-87, 2015. http://dx.doi.org/10.15698/mic2015.03.193
- R.J. Braun, and H. Zischka, "Mechanisms of Cdc48/VCP-mediated cell death — from yeast apoptosis to human disease", Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, vol. 1783, pp. 1418-1435, 2008. http://dx.doi.org/10.1016/j.bbamcr.2008.01.015
- Q. Xu, and J.C. Reed, "Bax inhibitor-1, a mammalian apoptosis suppressor identified by functional screening in yeast.", Molecular cell, 1998. http://www.ncbi.nlm.nih.gov/pubmed/9660918
- F. Madeo, E. Herker, C. Maldener, S. Wissing, S. Lächelt, M. Herlan, M. Fehr, K. Lauber, S.J. Sigrist, S. Wesselborg, and K.U. Fröhlich, "A caspase-related protease regulates apoptosis in yeast.", Molecular cell, 2002. http://www.ncbi.nlm.nih.gov/pubmed/11983181
- J.F. Sundström, A. Vaculova, A.P. Smertenko, E.I. Savenkov, A. Golovko, E. Minina, B.S. Tiwari, S. Rodriguez-Nieto, A.A. Zamyatnin, T. Välineva, J. Saarikettu, M.J. Frilander, M.F. Suarez, A. Zavialov, U. Ståhl, P.J. Hussey, O. Silvennoinen, E. Sundberg, B. Zhivotovsky, and P.V. Bozhkov, "Tudor staphylococcal nuclease is an evolutionarily conserved component of the programmed cell death degradome", Nature Cell Biology, vol. 11, pp. 1347-1354, 2009. http://dx.doi.org/10.1038/ncb1979
- D. Carmona-Gutierrez, K. Fröhlich, G. Kroemer, and F. Madeo, "Metacaspases are caspases. Doubt no more", Cell Death & Differentiation, vol. 17, pp. 377-378, 2010. http://dx.doi.org/10.1038/cdd.2009.198
- C.S.F. Oliveira, H. Pereira, S. Alves, L. Castro, F. Baltazar, S.R. Chaves, A. Preto, and M. Côrte-Real, "Cathepsin D protects colorectal cancer cells from acetate-induced apoptosis through autophagy-independent degradation of damaged mitochondria", Cell Death & Disease, vol. 6, pp. e1788-e1788, 2015. http://dx.doi.org/10.1038/cddis.2015.157
- C. Pereira, S. Chaves, S. Alves, B. Salin, N. Camougrand, S. Manon, M.J. Sousa, and M. Côrte-Real, "Mitochondrial degradation in acetic acid-induced yeast apoptosis: the role of Pep4 and the ADP/ATP carrier", Molecular Microbiology, vol. 76, pp. 1398-1410, 2010. http://dx.doi.org/10.1111/j.1365-2958.2010.07122.x
- A.A. Cooper, A.D. Gitler, A. Cashikar, C.M. Haynes, K.J. Hill, B. Bhullar, K. Liu, K. Xu, K.E. Strathearn, F. Liu, S. Cao, K.A. Caldwell, G.A. Caldwell, G. Marsischky, R.D. Kolodner, J. LaBaer, J. Rochet, N.M. Bonini, and S. Lindquist, "α-Synuclein Blocks ER-Golgi Traffic and Rab1 Rescues Neuron Loss in Parkinson's Models", Science, vol. 313, pp. 324-328, 2006. http://dx.doi.org/10.1126/science.1129462
- E. Herker, H. Jungwirth, K.A. Lehmann, C. Maldener, K. Fröhlich, S. Wissing, S. Büttner, M. Fehr, S. Sigrist, and F. Madeo, "Chronological aging leads to apoptosis in yeast", The Journal of Cell Biology, vol. 164, pp. 501-507, 2004. http://dx.doi.org/10.1083/jcb.200310014
- S. Büttner, T. Eisenberg, E. Herker, D. Carmona-Gutierrez, G. Kroemer, and F. Madeo, "Why yeast cells can undergo apoptosis: death in times of peace, love, and war", The Journal of Cell Biology, vol. 175, pp. 521-525, 2006. http://dx.doi.org/10.1083/jcb.200608098
- Z. Palková, B. Janderová, J. Gabriel, B. Zikánová, M. Pospíŝek, and J. Forstová, "Ammonia mediates communication between yeast colonies", Nature, vol. 390, pp. 532-536, 1997. http://dx.doi.org/10.1038/37398
- M. Čáp, L. Váchová, and Z. Palková, "Yeast Colony Survival Depends on Metabolic Adaptation and Cell Differentiation Rather Than on Stress Defense", Journal of Biological Chemistry, vol. 284, pp. 32572-32581, 2009. http://dx.doi.org/10.1074/jbc.M109.022871
- L. Váchová, and Z. Palková, "Physiological regulation of yeast cell death in multicellular colonies is triggered by ammonia", The Journal of Cell Biology, vol. 169, pp. 711-717, 2005. http://dx.doi.org/10.1083/jcb.200410064
- D. Knorre, S. Sokolov, A. Zyrina, and F. Severin, "How do yeast sense mitochondrial dysfunction?", Microbial Cell, vol. 3, pp. 532-539, 2016. http://dx.doi.org/10.15698/mic2016.11.537
- F.F. Severin, and A.A. Hyman, "Pheromone induces programmed cell death in S. cerevisiae.", Current biology : CB, 2002. http://www.ncbi.nlm.nih.gov/pubmed/11937036
- J. Reiter, E. Herker, F. Madeo, and M.J. Schmitt, "Viral killer toxins induce caspase-mediated apoptosis in yeast", The Journal of Cell Biology, vol. 168, pp. 353-358, 2005. http://dx.doi.org/10.1083/jcb.200408071
- P. Fabrizio, L. Battistella, R. Vardavas, C. Gattazzo, L. Liou, A. Diaspro, J.W. Dossen, E.B. Gralla, and V.D. Longo, "Superoxide is a mediator of an altruistic aging program in Saccharomyces cerevisiae ", The Journal of Cell Biology, vol. 166, pp. 1055-1067, 2004. http://dx.doi.org/10.1083/jcb.200404002
- R. Klassen, R. Schaffrath, P. Buzzini, and P.F. Ganter, "Antagonistic Interactions and Killer Yeasts", Yeasts in Natural Ecosystems: Ecology, pp. 229-275, 2017. http://dx.doi.org/10.1007/978-3-319-61575-2_9
- R. Klassen, and F. Meinhardt, "Induction of DNA damage and apoptosis in Saccharomyces cerevisiae by a yeast killer toxin", Cellular Microbiology, vol. 7, pp. 393-401, 2005. http://dx.doi.org/10.1111/j.1462-5822.2004.00469.x
- D. Carmona-Gutierrez, G. Kroemer, and F. Madeo, "When Death Was Young: An Ancestral Apoptotic Network in Bacteria", Molecular Cell, vol. 46, pp. 552-554, 2012. http://dx.doi.org/10.1016/j.molcel.2012.05.032
- D. Dwyer, D. Camacho, M. Kohanski, J. Callura, and J. Collins, "Antibiotic-Induced Bacterial Cell Death Exhibits Physiological and Biochemical Hallmarks of Apoptosis", Molecular Cell, vol. 46, pp. 561-572, 2012. http://dx.doi.org/10.1016/j.molcel.2012.04.027
- F. Sow, M. Nyonda, A. Bienvenu, and S. Picot, "Wanted Plasmodium falciparum, dead or alive", Microbial Cell, vol. 2, pp. 219-224, 2015. http://dx.doi.org/10.15698/mic2015.07.211
- A. Debrabant, N. Lee, S. Bertholet, R. Duncan, and H.L. Nakhasi, "Programmed cell death in trypanosomatids and other unicellular organisms.", International journal for parasitology, 2003. http://www.ncbi.nlm.nih.gov/pubmed/12670511
- E. Taylor-Brown, and H. Hurd, "The first suicides: a legacy inherited by parasitic protozoans from prokaryote ancestors", Parasites & Vectors, vol. 6, 2013. http://dx.doi.org/10.1186/1756-3305-6-108
- J. Winderickx, and P. Ludovico, "Microbial Programmed Necrosis: The Cost of Conflicts Between Stress and Metabolism", Necrotic Cell Death, pp. 253-274, 2014. http://dx.doi.org/10.1007/978-1-4614-8220-8_14
- L. Galluzzi, S.A. Aaronson, J. Abrams, E.S. Alnemri, D.W. Andrews, E.H. Baehrecke, N.G. Bazan, M.V. Blagosklonny, K. Blomgren, C. Borner, D.E. Bredesen, C. Brenner, M. Castedo, J.A. Cidlowski, A. Ciechanover, G.M. Cohen, V. De Laurenzi, R. De Maria, M. Deshmukh, B.D. Dynlacht, W.S. El-Deiry, R.A. Flavell, S. Fulda, C. Garrido, P. Golstein, M. Gougeon, D.R. Green, H. Gronemeyer, G. Hajnóczky, J.M. Hardwick, M.O. Hengartner, H. Ichijo, M. Jäättelä, O. Kepp, A. Kimchi, D.J. Klionsky, R.A. Knight, S. Kornbluth, S. Kumar, B. Levine, S.A. Lipton, E. Lugli, F. Madeo, W. Malorni, J. Marine, S.J. Martin, J.P. Medema, P. Mehlen, G. Melino, U.M. Moll, E. Morselli, S. Nagata, D.W. Nicholson, P. Nicotera, G. Nuñez, M. Oren, J. Penninger, S. Pervaiz, M.E. Peter, M. Piacentini, J.H.M. Prehn, H. Puthalakath, G.A. Rabinovich, R. Rizzuto, C.M.P. Rodrigues, D.C. Rubinsztein, T. Rudel, L. Scorrano, H. Simon, H. Steller, J. Tschopp, Y. Tsujimoto, P. Vandenabeele, I. Vitale, K.H. Vousden, R.J. Youle, J. Yuan, B. Zhivotovsky, and G. Kroemer, "Guidelines for the use and interpretation of assays for monitoring cell death in higher eukaryotes", Cell Death & Differentiation, vol. 16, pp. 1093-1107, 2009. http://dx.doi.org/10.1038/cdd.2009.44
- L. Galluzzi, J.M. Bravo-San Pedro, I. Vitale, S.A. Aaronson, J.M. Abrams, D. Adam, E.S. Alnemri, L. Altucci, D. Andrews, M. Annicchiarico-Petruzzelli, E.H. Baehrecke, N.G. Bazan, M.J. Bertrand, K. Bianchi, M.V. Blagosklonny, K. Blomgren, C. Borner, D.E. Bredesen, C. Brenner, M. Campanella, E. Candi, F. Cecconi, F.K. Chan, N.S. Chandel, E.H. Cheng, J.E. Chipuk, J.A. Cidlowski, A. Ciechanover, T.M. Dawson, V.L. Dawson, V. De Laurenzi, R. De Maria, K. Debatin, N. Di Daniele, V.M. Dixit, B.D. Dynlacht, W.S. El-Deiry, G.M. Fimia, R.A. Flavell, S. Fulda, C. Garrido, M. Gougeon, D.R. Green, H. Gronemeyer, G. Hajnoczky, J.M. Hardwick, M.O. Hengartner, H. Ichijo, B. Joseph, P.J. Jost, T. Kaufmann, O. Kepp, D.J. Klionsky, R.A. Knight, S. Kumar, J.J. Lemasters, B. Levine, A. Linkermann, S.A. Lipton, R.A. Lockshin, C. López-Otín, E. Lugli, F. Madeo, W. Malorni, J. Marine, S.J. Martin, J. Martinou, J.P. Medema, P. Meier, S. Melino, N. Mizushima, U. Moll, C. Muñoz-Pinedo, G. Nuñez, A. Oberst, T. Panaretakis, J.M. Penninger, M.E. Peter, M. Piacentini, P. Pinton, J.H. Prehn, H. Puthalakath, G.A. Rabinovich, K.S. Ravichandran, R. Rizzuto, C.M. Rodrigues, D.C. Rubinsztein, T. Rudel, Y. Shi, H. Simon, B.R. Stockwell, G. Szabadkai, S.W. Tait, H.L. Tang, N. Tavernarakis, Y. Tsujimoto, T. Vanden Berghe, P. Vandenabeele, A. Villunger, E.F. Wagner, H. Walczak, E. White, W.G. Wood, J. Yuan, Z. Zakeri, B. Zhivotovsky, G. Melino, and G. Kroemer, "Essential versus accessory aspects of cell death: recommendations of the NCCD 2015", Cell Death & Differentiation, vol. 22, pp. 58-73, 2014. http://dx.doi.org/10.1038/cdd.2014.137
- L. Galluzzi, I. Vitale, J.M. Abrams, E.S. Alnemri, E.H. Baehrecke, M.V. Blagosklonny, T.M. Dawson, V.L. Dawson, W.S. El-Deiry, S. Fulda, E. Gottlieb, D.R. Green, M.O. Hengartner, O. Kepp, R.A. Knight, S. Kumar, S.A. Lipton, X. Lu, F. Madeo, W. Malorni, P. Mehlen, G. Nuñez, M.E. Peter, M. Piacentini, D.C. Rubinsztein, Y. Shi, H. Simon, P. Vandenabeele, E. White, J. Yuan, B. Zhivotovsky, G. Melino, and G. Kroemer, "Molecular definitions of cell death subroutines: recommendations of the Nomenclature Committee on Cell Death 2012", Cell Death & Differentiation, vol. 19, pp. 107-120, 2011. http://dx.doi.org/10.1038/cdd.2011.96
- L. Galluzzi, I. Vitale, and E. al, "Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death 2018.", Cell Death Differ in press., 2018.
- S. Büttner, T. Eisenberg, D. Carmona-Gutierrez, D. Ruli, H. Knauer, C. Ruckenstuhl, C. Sigrist, S. Wissing, M. Kollroser, K. Fröhlich, S. Sigrist, and F. Madeo, "Endonuclease G Regulates Budding Yeast Life and Death", Molecular Cell, vol. 25, pp. 233-246, 2007. http://dx.doi.org/10.1016/j.molcel.2006.12.021
- D. Deere, J. Shen, G. Vesey, P. Bell, P. Bissinger, and D. Veal, "Flow cytometry and cell sorting for yeast viability assessment and cell selection.", Yeast (Chichester, England), 1998. http://www.ncbi.nlm.nih.gov/pubmed/9483803
- M.G. Mirisola, R.J. Braun, and D. Petranovic, "Approaches to study yeast cell aging and death", FEMS Yeast Research, vol. 14, pp. 109-118, 2013. http://dx.doi.org/10.1111/1567-1364.12112
- E. Keyhani, S. Khavari‐Nejad, J. Keyhani, and F. Attar, "Acriflavine‐Mediated Apoptosis and Necrosis in Yeast Candida utilis", Annals of the New York Academy of Sciences, vol. 1171, pp. 284-291, 2009. http://dx.doi.org/10.1111/j.1749-6632.2009.04682.x
- M. Kwolek-Mirek, and R. Zadrag-Tecza, "Comparison of methods used for assessing the viability and vitality of yeast cells", FEMS Yeast Research, pp. n/a-n/a, 2014. http://dx.doi.org/10.1111/1567-1364.12202
- J. Kucsera, K. Yarita, and K. Takeo, "Simple detection method for distinguishing dead and living yeast colonies", Journal of Microbiological Methods, vol. 41, pp. 19-21, 2000. http://dx.doi.org/10.1016/S0167-7012(00)00136-6
- H.M. Davey, and P. Hexley, "Red but not dead? Membranes of stressed Saccharomyces cerevisiae are permeable to propidium iodide", Environmental Microbiology, vol. 13, pp. 163-171, 2011. http://dx.doi.org/10.1111/j.1462-2920.2010.02317.x
- T. Eisenberg, D. Carmona-Gutierrez, S. Büttner, N. Tavernarakis, and F. Madeo, "Necrosis in yeast", Apoptosis, vol. 15, pp. 257-268, 2010. http://dx.doi.org/10.1007/s10495-009-0453-4
- D. Carmona-Gutiérrez, M.A. Bauer, J. Ring, H. Knauer, T. Eisenberg, S. Büttner, C. Ruckenstuhl, A. Reisenbichler, C. Magnes, G.N. Rechberger, R. Birner-Gruenberger, H. Jungwirth, K. Fröhlich, F. Sinner, G. Kroemer, and F. Madeo, "The propeptide of yeast cathepsin D inhibits programmed necrosis", Cell Death & Disease, vol. 2, pp. e161-e161, 2011. http://dx.doi.org/10.1038/cddis.2011.43
- C. Ruckenstuhl, C. Netzberger, I. Entfellner, D. Carmona-Gutierrez, T. Kickenweiz, S. Stekovic, C. Gleixner, C. Schmid, L. Klug, A.G. Sorgo, T. Eisenberg, S. Büttner, G. Mariño, R. Koziel, P. Jansen-Dürr, K. Fröhlich, G. Kroemer, and F. Madeo, "Lifespan Extension by Methionine Restriction Requires Autophagy-Dependent Vacuolar Acidification", PLoS Genetics, vol. 10, pp. e1004347, 2014. http://dx.doi.org/10.1371/journal.pgen.1004347
- V. Palermo, C. Falcone, and C. Mazzoni, "Apoptosis and aging in mitochondrial morphology mutants of S. cerevisiae.", Folia microbiologica, 2007. http://www.ncbi.nlm.nih.gov/pubmed/18298044
-
V. Palermo, C. Falcone, M. Calvani, and C. Mazzoni, "Acetyl
‐l‐ carnitine protects yeast cells from apoptosis and aging and inhibits mitochondrial fission", Aging Cell, vol. 9, pp. 570-579, 2010. http://dx.doi.org/10.1111/j.1474-9726.2010.00587.x - P.P. Jung, N. Christian, D.P. Kay, A. Skupin, and C.L. Linster, "Protocols and Programs for High-Throughput Growth and Aging Phenotyping in Yeast", PLOS ONE, vol. 10, pp. e0119807, 2015. http://dx.doi.org/10.1371/journal.pone.0119807
- R.W. Powers, M. Kaeberlein, S.D. Caldwell, B.K. Kennedy, and S. Fields, "Extension of chronological life span in yeast by decreased TOR pathway signaling", Genes & Development, vol. 20, pp. 174-184, 2006. http://dx.doi.org/10.1101/gad.1381406
- F. Giorgini, P. Guidetti, Q. Nguyen, S.C. Bennett, and P.J. Muchowski, "A genomic screen in yeast implicates kynurenine 3-monooxygenase as a therapeutic target for Huntington disease", Nature Genetics, vol. 37, pp. 526-531, 2005. http://dx.doi.org/10.1038/ng1542
- X. Teng, and J.M. Hardwick, "Quantification of Genetically Controlled Cell Death in Budding Yeast", Methods in Molecular Biology, pp. 161-170, 2013. http://dx.doi.org/10.1007/978-1-62703-383-1_12
- P. Breeuwer, J.L. Drocourt, N. Bunschoten, M.H. Zwietering, F.M. Rombouts, and T. Abee, "Characterization of uptake and hydrolysis of fluorescein diacetate and carboxyfluorescein diacetate by intracellular esterases in Saccharomyces cerevisiae, which result in accumulation of fluorescent product.", Applied and environmental microbiology, 1995. http://www.ncbi.nlm.nih.gov/pubmed/7747975
- E.M. Czekanska, "Assessment of Cell Proliferation with Resazurin-Based Fluorescent Dye", Methods in Molecular Biology, pp. 27-32, 2011. http://dx.doi.org/10.1007/978-1-61779-108-6_5
- Y. Fannjiang, W. Cheng, S.J. Lee, B. Qi, J. Pevsner, J.M. McCaffery, R.B. Hill, G. Basañez, and J.M. Hardwick, "Mitochondrial fission proteins regulate programmed cell death in yeast", Genes & Development, vol. 18, pp. 2785-2797, 2004. http://dx.doi.org/10.1101/gad.1247904
- X. Teng, and J.M. Hardwick, "Reliable Method for Detection of Programmed Cell Death in Yeast", Methods in Molecular Biology, pp. 335-342, 2009. http://dx.doi.org/10.1007/978-1-60327-017-5_23
- D.M. Kuhn, M. Balkis, J. Chandra, P.K. Mukherjee, and M.A. Ghannoum, "Uses and Limitations of the XTT Assay in Studies of Candida Growth and Metabolism", Journal of Clinical Microbiology, vol. 41, pp. 506-508, 2003. http://dx.doi.org/10.1128/JCM.41.1.506-508.2003
- P. Bapat, S.K. Nandy, P. Wangikar, and K. Venkatesh, "Quantification of metabolically active biomass using Methylene Blue dye Reduction Test (MBRT): Measurement of CFU in about 200 s", Journal of Microbiological Methods, vol. 65, pp. 107-116, 2006. http://dx.doi.org/10.1016/j.mimet.2005.06.010
- T. Noda, "Chapter 2 Viability Assays to Monitor Yeast Autophagy", Methods in Enzymology, pp. 27-32, 2008. http://dx.doi.org/10.1016/S0076-6879(08)03202-3
- S. Anséhn, and L. Nilsson, "Direct membrane-damaging effect of ketoconazole and tioconazole on Candida albicans demonstrated by bioluminescent assay of ATP", Antimicrobial Agents and Chemotherapy, vol. 26, pp. 22-25, 1984. http://dx.doi.org/10.1128/AAC.26.1.22
- P. Ludovico, M. Côrte-Real, and F. Sansonetty, "Assessment of mitochondrial membrane potential in yeast cell populations by flow cytometry", Microbiology, vol. 147, pp. 3335-3343, 2001. http://dx.doi.org/10.1099/00221287-147-12-3335
- C. Cottet‐Rousselle, X. Ronot, X. Leverve, and J. Mayol, "Cytometric assessment of mitochondria using fluorescent probes", Cytometry Part A, vol. 79A, pp. 405-425, 2011. http://dx.doi.org/10.1002/cyto.a.21061
- G.G. Perrone, S. Tan, and I.W. Dawes, "Reactive oxygen species and yeast apoptosis", Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, vol. 1783, pp. 1354-1368, 2008. http://dx.doi.org/10.1016/j.bbamcr.2008.01.023
- Y.T. Lam, M.T. Aung-Htut, Y.L. Lim, H. Yang, and I.W. Dawes, "Changes in reactive oxygen species begin early during replicative aging of Saccharomyces cerevisiae cells", Free Radical Biology and Medicine, vol. 50, pp. 963-970, 2011. http://dx.doi.org/10.1016/j.freeradbiomed.2011.01.013
- P. Laun, A. Pichova, F. Madeo, J. Fuchs, A. Ellinger, S. Kohlwein, I. Dawes, K.U. Fröhlich, and M. Breitenbach, "Aged mother cells of Saccharomyces cerevisiae show markers of oxidative stress and apoptosis.", Molecular microbiology, 2001. http://www.ncbi.nlm.nih.gov/pubmed/11251834
- F. Madeo, E. Fröhlich, M. Ligr, M. Grey, S.J. Sigrist, D.H. Wolf, and K.U. Fröhlich, "Oxygen stress: a regulator of apoptosis in yeast.", The Journal of cell biology, 1999. http://www.ncbi.nlm.nih.gov/pubmed/10330404
- B. Halliwell, and M. Whiteman, "Measuring reactive species and oxidative damagein vivoand in cell culture: how should you do it and what do the results mean?", British Journal of Pharmacology, vol. 142, pp. 231-255, 2004. http://dx.doi.org/10.1038/sj.bjp.0705776
- R.R. Nazarewicz, A. Bikineyeva, and S.I. Dikalov, "Rapid and Specific Measurements of Superoxide Using Fluorescence Spectroscopy", SLAS Discovery, vol. 18, pp. 498-503, 2013. http://dx.doi.org/10.1177/1087057112468765
- H. Zhao, J. Joseph, H.M. Fales, E.A. Sokoloski, R.L. Levine, J. Vasquez-Vivar, and B. Kalyanaraman, "Detection and characterization of the product of hydroethidine and intracellular superoxide by HPLC and limitations of fluorescence", Proceedings of the National Academy of Sciences, vol. 102, pp. 5727-5732, 2005. http://dx.doi.org/10.1073/pnas.0501719102
- T. Finkel, "Signal transduction by reactive oxygen species", Journal of Cell Biology, vol. 194, pp. 7-15, 2011. http://dx.doi.org/10.1083/jcb.201102095
- J.H.M. Schippers, H.M. Nguyen, D. Lu, R. Schmidt, and B. Mueller-Roeber, "ROS homeostasis during development: an evolutionary conserved strategy", Cellular and Molecular Life Sciences, vol. 69, pp. 3245-3257, 2012. http://dx.doi.org/10.1007/s00018-012-1092-4
- L. Sena, and N. Chandel, "Physiological Roles of Mitochondrial Reactive Oxygen Species", Molecular Cell, vol. 48, pp. 158-167, 2012. http://dx.doi.org/10.1016/j.molcel.2012.09.025
- M. Rinnerthaler, S. Büttner, P. Laun, G. Heeren, T.K. Felder, H. Klinger, M. Weinberger, K. Stolze, T. Grousl, J. Hasek, O. Benada, I. Frydlova, A. Klocker, B. Simon-Nobbe, B. Jansko, H. Breitenbach-Koller, T. Eisenberg, C.W. Gourlay, F. Madeo, W.C. Burhans, and M. Breitenbach, "Yno1p/Aim14p, a NADPH-oxidase ortholog, controls extramitochondrial reactive oxygen species generation, apoptosis, and actin cable formation in yeast", Proceedings of the National Academy of Sciences, vol. 109, pp. 8658-8663, 2012. http://dx.doi.org/10.1073/pnas.1201629109
- M. Chi, and K.D. Craven, "RacA-Mediated ROS Signaling Is Required for Polarized Cell Differentiation in Conidiogenesis of Aspergillus fumigatus", PLOS ONE, vol. 11, pp. e0149548, 2016. http://dx.doi.org/10.1371/journal.pone.0149548
- A.M. Day, and E.A. Veal, "Hydrogen Peroxide-sensitive Cysteines in the Sty1 MAPK Regulate the Transcriptional Response to Oxidative Stress", Journal of Biological Chemistry, vol. 285, pp. 7505-7516, 2010. http://dx.doi.org/10.1074/jbc.M109.040840
- B.J. Niles, and T. Powers, "TOR complex 2–Ypk1 signaling regulates actin polarization via reactive oxygen species", Molecular Biology of the Cell, vol. 25, pp. 3962-3972, 2014. http://dx.doi.org/10.1091/mbc.E14-06-1122
- G. Shadel, . , and . , "Live longer on MARS: a yeast paradigm of mitochondrial adaptive ROS signaling in aging", Microbial Cell, vol. 1, pp. 140-144, 2014. http://dx.doi.org/10.15698/mic2014.05.143
- A. Mesquita, M. Weinberger, A. Silva, B. Sampaio-Marques, B. Almeida, C. Leão, V. Costa, F. Rodrigues, W.C. Burhans, and P. Ludovico, "Caloric restriction or catalase inactivation extends yeast chronological lifespan by inducing H 2 O 2 and superoxide dismutase activity", Proceedings of the National Academy of Sciences, vol. 107, pp. 15123-15128, 2010. http://dx.doi.org/10.1073/pnas.1004432107
- P. Ludovico, and W.C. Burhans, "Reactive oxygen species, ageing and the hormesis police", FEMS Yeast Research, vol. 14, pp. 33-39, 2013. http://dx.doi.org/10.1111/1567-1364.12070
- S.D. Postnikoff, J.E. Johnson, and J.K. Tyler, "The integrated stress response in budding yeast lifespan extension", Microbial Cell, vol. 4, pp. 368-375, 2017. http://dx.doi.org/10.15698/mic2017.11.597
- S. Zelenay, and C. Reis e Sousa, "Adaptive immunity after cell death", Trends in Immunology, vol. 34, pp. 329-335, 2013. http://dx.doi.org/10.1016/j.it.2013.03.005
- Q. Zhang, M. Raoof, Y. Chen, Y. Sumi, T. Sursal, W. Junger, K. Brohi, K. Itagaki, and C.J. Hauser, "Circulating mitochondrial DAMPs cause inflammatory responses to injury", Nature, vol. 464, pp. 104-107, 2010. http://dx.doi.org/10.1038/nature08780
- A.D. Garg, L. Galluzzi, L. Apetoh, T. Baert, R.B. Birge, J.M. Bravo-San Pedro, K. Breckpot, D. Brough, R. Chaurio, M. Cirone, A. Coosemans, P.G. Coulie, D. De Ruysscher, L. Dini, P. de Witte, A.M. Dudek-Peric, A. Faggioni, J. Fucikova, U.S. Gaipl, J. Golab, M. Gougeon, M.R. Hamblin, A. Hemminki, M. Herrmann, J.W. Hodge, O. Kepp, G. Kroemer, D.V. Krysko, W.G. Land, F. Madeo, A.A. Manfredi, S.R. Mattarollo, C. Maueroder, N. Merendino, G. Multhoff, T. Pabst, J. Ricci, C. Riganti, E. Romano, N. Rufo, M.J. Smyth, J. Sonnemann, R. Spisek, J. Stagg, E. Vacchelli, P. Vandenabeele, L. Vandenberk, B.J. Van den Eynde, S. Van Gool, F. Velotti, L. Zitvogel, and P. Agostinis, "Molecular and Translational Classifications of DAMPs in Immunogenic Cell Death", Frontiers in Immunology, vol. 6, 2015. http://dx.doi.org/10.3389/fimmu.2015.00588
- O. Kepp, L. Senovilla, I. Vitale, E. Vacchelli, S. Adjemian, P. Agostinis, L. Apetoh, F. Aranda, V. Barnaba, N. Bloy, L. Bracci, K. Breckpot, D. Brough, A. Buqué, M.G. Castro, M. Cirone, M.I. Colombo, I. Cremer, S. Demaria, L. Dini, A.G. Eliopoulos, A. Faggioni, S.C. Formenti, J. Fučíková, L. Gabriele, U.S. Gaipl, J. Galon, A. Garg, F. Ghiringhelli, N.A. Giese, Z.S. Guo, A. Hemminki, M. Herrmann, J.W. Hodge, S. Holdenrieder, J. Honeychurch, H. Hu, X. Huang, T.M. Illidge, K. Kono, M. Korbelik, D.V. Krysko, S. Loi, P.R. Lowenstein, E. Lugli, Y. Ma, F. Madeo, A.A. Manfredi, I. Martins, D. Mavilio, L. Menger, N. Merendino, M. Michaud, G. Mignot, K.L. Mossman, G. Multhoff, R. Oehler, F. Palombo, T. Panaretakis, J. Pol, E. Proietti, J. Ricci, C. Riganti, P. Rovere-Querini, A. Rubartelli, A. Sistigu, M.J. Smyth, J. Sonnemann, R. Spisek, J. Stagg, A.Q. Sukkurwala, E. Tartour, A. Thorburn, S.H. Thorne, P. Vandenabeele, F. Velotti, S.T. Workenhe, H. Yang, W. Zong, L. Zitvogel, G. Kroemer, and L. Galluzzi, "Consensus guidelines for the detection of immunogenic cell death", OncoImmunology, vol. 3, pp. e955691, 2014. http://dx.doi.org/10.4161/21624011.2014.955691
- L. Galluzzi, A. Buqué, O. Kepp, L. Zitvogel, and G. Kroemer, "Immunogenic cell death in cancer and infectious disease", Nature Reviews Immunology, vol. 17, pp. 97-111, 2016. http://dx.doi.org/10.1038/nri.2016.107
- A.R.D. Delbridge, L.J. Valente, and A. Strasser, "The Role of the Apoptotic Machinery in Tumor Suppression", Cold Spring Harbor Perspectives in Biology, vol. 4, pp. a008789-a008789, 2012. http://dx.doi.org/10.1101/cshperspect.a008789
- Y. Fuchs, and H. Steller, "Programmed Cell Death in Animal Development and Disease", Cell, vol. 147, pp. 742-758, 2011. http://dx.doi.org/10.1016/j.cell.2011.10.033
- G. Lettre, and M.O. Hengartner, "Developmental apoptosis in C. elegans: a complex CEDnario", Nature Reviews Molecular Cell Biology, vol. 7, pp. 97-108, 2006. http://dx.doi.org/10.1038/nrm1836
- R.C. Taylor, S.P. Cullen, and S.J. Martin, "Apoptosis: controlled demolition at the cellular level", Nature Reviews Molecular Cell Biology, vol. 9, pp. 231-241, 2008. http://dx.doi.org/10.1038/nrm2312
- M.D. Eastwood, and M.D. Meneghini, "Developmental Coordination of Gamete Differentiation with Programmed Cell Death in Sporulating Yeast", Eukaryotic Cell, vol. 14, pp. 858-867, 2015. http://dx.doi.org/10.1128/EC.00068-15
- M. Eastwood, S. Cheung, K. Lee, J. Moffat, and M. Meneghini, "Developmentally Programmed Nuclear Destruction during Yeast Gametogenesis", Developmental Cell, vol. 23, pp. 35-44, 2012. http://dx.doi.org/10.1016/j.devcel.2012.05.005
- M. Čáp, L. Štěpánek, K. Harant, L. Váchová, and Z. Palková, "Cell Differentiation within a Yeast Colony: Metabolic and Regulatory Parallels with a Tumor-Affected Organism", Molecular Cell, vol. 46, pp. 436-448, 2012. http://dx.doi.org/10.1016/j.molcel.2012.04.001
- Z. Palková, and L. Váchová, "Yeast cell differentiation: Lessons from pathogenic and non-pathogenic yeasts", Seminars in Cell & Developmental Biology, vol. 57, pp. 110-119, 2016. http://dx.doi.org/10.1016/j.semcdb.2016.04.006
- L. Váchová, and Z. Palková, "Caspases in yeast apoptosis-like death: facts and artefacts", FEMS Yeast Research, vol. 7, pp. 12-21, 2007. http://dx.doi.org/10.1111/j.1567-1364.2006.00137.x
- W. Cheng, X. Teng, H.K. Park, C.M. Tucker, M.J. Dunham, and J.M. Hardwick, "Fis1 deficiency selects for compensatory mutations responsible for cell death and growth control defects", Cell Death & Differentiation, vol. 15, pp. 1838-1846, 2008. http://dx.doi.org/10.1038/cdd.2008.117
- G. Kroemer, L. Galluzzi, P. Vandenabeele, J. Abrams, E.S. Alnemri, E.H. Baehrecke, M.V. Blagosklonny, W.S. El-Deiry, P. Golstein, D.R. Green, M. Hengartner, R.A. Knight, S. Kumar, S.A. Lipton, W. Malorni, G. Nuñez, M.E. Peter, J. Tschopp, J. Yuan, M. Piacentini, B. Zhivotovsky, and G. Melino, "Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009", Cell Death & Differentiation, vol. 16, pp. 3-11, 2008. http://dx.doi.org/10.1038/cdd.2008.150
- D.V. Krysko, T.V. Berghe, E. Parthoens, K. D'Herde, and P. Vandenabeele, "Chapter 16 Methods for Distinguishing Apoptotic from Necrotic Cells and Measuring Their Clearance", Programmed Cell Death,General Principles forStudying Cell Death, Part A, pp. 307-341, 2008. http://dx.doi.org/10.1016/S0076-6879(08)01416-X
- S.J. Martin, C.P. Reutelingsperger, A.J. McGahon, J.A. Rader, R.C. van Schie, D.M. LaFace, and D.R. Green, "Early redistribution of plasma membrane phosphatidylserine is a general feature of apoptosis regardless of the initiating stimulus: inhibition by overexpression of Bcl-2 and Abl.", The Journal of experimental medicine, vol. 182, pp. 1545-1556, 1995. http://dx.doi.org/10.1084/jem.182.5.1545
- M.A. Mellén, E.J. de la Rosa, and P. Boya, "Autophagy is not universally required for phosphatidyl-serine exposure and apoptotic cell engulfment during neural development", Autophagy, vol. 5, pp. 964-972, 2009. http://dx.doi.org/10.4161/auto.5.7.9292
- K. Segawa, S. Kurata, Y. Yanagihashi, T.R. Brummelkamp, F. Matsuda, and S. Nagata, "Caspase-mediated cleavage of phospholipid flippase for apoptotic phosphatidylserine exposure", Science, vol. 344, pp. 1164-1168, 2014. http://dx.doi.org/10.1126/science.1252809
- T. Eisenberg, H. Knauer, A. Schauer, S. Büttner, C. Ruckenstuhl, D. Carmona-Gutierrez, J. Ring, S. Schroeder, C. Magnes, L. Antonacci, H. Fussi, L. Deszcz, R. Hartl, E. Schraml, A. Criollo, E. Megalou, D. Weiskopf, P. Laun, G. Heeren, M. Breitenbach, B. Grubeck-Loebenstein, E. Herker, B. Fahrenkrog, K. Fröhlich, F. Sinner, N. Tavernarakis, N. Minois, G. Kroemer, and F. Madeo, "Induction of autophagy by spermidine promotes longevity", Nature Cell Biology, vol. 11, pp. 1305-1314, 2009. http://dx.doi.org/10.1038/ncb1975
- P. Golstein, and G. Kroemer, "Cell death by necrosis: towards a molecular definition", Trends in Biochemical Sciences, vol. 32, pp. 37-43, 2007. http://dx.doi.org/10.1016/j.tibs.2006.11.001
- M.T. Silva, "Secondary necrosis: The natural outcome of the complete apoptotic program", FEBS Letters, vol. 584, pp. 4491-4499, 2010. http://dx.doi.org/10.1016/j.febslet.2010.10.046
- L. Galluzzi, and G. Kroemer, "Secondary Necrosis: Accidental No More", Trends in Cancer, vol. 3, pp. 1-2, 2017. http://dx.doi.org/10.1016/j.trecan.2016.12.001
- C. Rogers, T. Fernandes-Alnemri, L. Mayes, D. Alnemri, G. Cingolani, and E.S. Alnemri, "Cleavage of DFNA5 by caspase-3 during apoptosis mediates progression to secondary necrotic/pyroptotic cell death", Nature Communications, vol. 8, 2017. http://dx.doi.org/10.1038/ncomms14128
- H. Nakaoka, "Live fast, die fast principle in a single cell of fission yeast", Microbial Cell, vol. 4, pp. 308-310, 2017. http://dx.doi.org/10.15698/mic2017.09.591
- M. Cabrera, D. Novarina, I.L. Rempel, L.M. Veenhoff, and M. Chang, "A simple microfluidic platform to study age-dependent protein abundance and localization changes in Saccharomyces cerevisiae", Microbial Cell, vol. 4, pp. 169-174, 2017. http://dx.doi.org/10.15698/mic2017.05.573
- H. Nakaoka, and Y. Wakamoto, "Aging, mortality, and the fast growth trade-off of Schizosaccharomyces pombe", PLOS Biology, vol. 15, pp. e2001109, 2017. http://dx.doi.org/10.1371/journal.pbio.2001109
- S.S. Lee, I.A. Vizcarra, D.H.E.W. Huberts, L.P. Lee, and M. Heinemann, "Whole lifespan microscopic observation of budding yeast aging through a microfluidic dissection platform", Proceedings of the National Academy of Sciences, vol. 109, pp. 4916-4920, 2012. http://dx.doi.org/10.1073/pnas.1113505109
- K.L. Chen, M.M. Crane, and M. Kaeberlein, "Microfluidic technologies for yeast replicative lifespan studies", Mechanisms of Ageing and Development, vol. 161, pp. 262-269, 2017. http://dx.doi.org/10.1016/j.mad.2016.03.009
- X. Wu, C. Molinaro, N. Johnson, and C.A. Casiano, "Secondary necrosis is a source of proteolytically modified forms of specific intracellular autoantigens: implications for systemic autoimmunity.", Arthritis and rheumatism, 2001. http://www.ncbi.nlm.nih.gov/pubmed/11710720
- N. Bresgen, I. Ohlenschläger, N. Wacht, S. Afazel, G. Ladurner, and P. Eckl, "Ferritin and FasL (CD95L) mediate density dependent apoptosis in primary rat hepatocytes", Journal of Cellular Physiology, vol. 217, pp. 800-808, 2008. http://dx.doi.org/10.1002/jcp.21555
- N.I. Walker, R.E. Bennett, and J.F.R. Kerr, "Cell death by apoptosis during involution of the lactating breast in mice and rats", American Journal of Anatomy, vol. 185, pp. 19-32, 1989. http://dx.doi.org/10.1002/aja.1001850104
- J. Ogasawara, R. Watanabe-Fukunaga, M. Adachi, A. Matsuzawa, T. Kasugai, Y. Kitamura, N. Itoh, T. Suda, and S. Nagata, "Lethal effect of the anti-Fas antibody in mice", Nature, vol. 364, pp. 806-809, 1993. http://dx.doi.org/10.1038/364806a0
- F. Madeo, E. Herker, C. Maldener, S. Wissing, S. Lächelt, M. Herlan, M. Fehr, K. Lauber, S.J. Sigrist, S. Wesselborg, and K. Fröhlich, "A Caspase-Related Protease Regulates Apoptosis in Yeast", Molecular Cell, vol. 9, pp. 911-917, 2002. http://dx.doi.org/10.1016/S1097-2765(02)00501-4
- B. Ansari, P.J. Coates, B.D. Greenstein, and P.A. Hall, "In situ end‐labelling detects DNA strand breaks in apoptosis and other physiological and pathological states", The Journal of Pathology, vol. 170, pp. 1-8, 1993. http://dx.doi.org/10.1002/path.1711700102
- V.V. Didenko, and P.J. Hornsby, "Presence of double-strand breaks with single-base 3' overhangs in cells undergoing apoptosis but not necrosis.", The Journal of cell biology, vol. 135, pp. 1369-1376, 1996. http://dx.doi.org/10.1083/jcb.135.5.1369
- M. Kanoh, G. Takemura, J. Misao, Y. Hayakawa, T. Aoyama, K. Nishigaki, T. Noda, T. Fujiwara, K. Fukuda, S. Minatoguchi, and H. Fujiwara, "Significance of myocytes with positive DNA in situ nick end-labeling (TUNEL) in hearts with dilated cardiomyopathy: not apoptosis but DNA repair.", Circulation, 1999. http://www.ncbi.nlm.nih.gov/pubmed/10351969
- M.M. Kockx, J. Muhring, M.W. Knaapen, and G.R. de Meyer, "RNA synthesis and splicing interferes with DNA in situ end labeling techniques used to detect apoptosis.", The American journal of pathology, 1998. http://www.ncbi.nlm.nih.gov/pubmed/9546348
- B.G. Kraupp, B. Ruttkay-Nedecky, H. Koudelka, K. Bukowska, W. Bursch, and R. Schulte-Hermann, "In situ detection of fragmented dna (tunel assay) fails to discriminate among apoptosis, necrosis, and autolytic cell death: A cautionary note", Hepatology, vol. 21, pp. 1465-1468, 1995. http://dx.doi.org/10.1002/hep.1840210534
- D.T. Loo, "In Situ Detection of Apoptosis by the TUNEL Assay: An Overview of Techniques", Methods in Molecular Biology, pp. 3-13, 2010. http://dx.doi.org/10.1007/978-1-60327-409-8_1
- G.F. Ribeiro, M. Côrte-Real, and B. Johansson, "Characterization of DNA Damage in Yeast Apoptosis Induced by Hydrogen Peroxide, Acetic Acid, and Hyperosmotic Shock", Molecular Biology of the Cell, vol. 17, pp. 4584-4591, 2006. http://dx.doi.org/10.1091/mbc.E06-05-0475
- B.D. Larsen, S. Rampalli, L.E. Burns, S. Brunette, F.J. Dilworth, and L.A. Megeney, "Caspase 3/caspase-activated DNase promote cell differentiation by inducing DNA strand breaks", Proceedings of the National Academy of Sciences, vol. 107, pp. 4230-4235, 2010. http://dx.doi.org/10.1073/pnas.0913089107
- R. Del Carratore, C. Della Croce, M. Simili, E. Taccini, M. Scavuzzo, and S. Sbrana, "Cell cycle and morphological alterations as indicative of apoptosis promoted by UV irradiation in S. cerevisiae.", Mutation research, 2002. http://www.ncbi.nlm.nih.gov/pubmed/11719103
- B. Almeida, B. Sampaio-Marques, J. Carvalho, M.T. Silva, C. Leão, F. Rodrigues, and P. Ludovico, "An atypical active cell death process underlies the fungicidal activity of ciclopirox olamine against the yeastSaccharomyces cerevisiae", FEMS Yeast Research, vol. 7, pp. 404-412, 2007. http://dx.doi.org/10.1111/j.1567-1364.2006.00188.x
- C.W. Gourlay, and K.R. Ayscough, "Identification of an upstream regulatory pathway controlling actin-mediated apoptosis in yeast", Journal of Cell Science, vol. 118, pp. 2119-2132, 2005. http://dx.doi.org/10.1242/jcs.02337
- P. Ludovico, F. Rodrigues, A. Almeida, M.T. Silva, A. Barrientos, and M. Côrte-Real, "CytochromecRelease and Mitochondria Involvement in Programmed Cell Death Induced by Acetic Acid inSaccharomyces cerevisiae", Molecular Biology of the Cell, vol. 13, pp. 2598-2606, 2002. http://dx.doi.org/10.1091/mbc.E01-12-0161
- J.R. Pringle, R.A. Preston, A.E. Adams, T. Stearns, D.G. Drubin, B.K. Haarer, and E.W. Jones, "Fluorescence microscopy methods for yeast.", Methods in cell biology, 1989. http://www.ncbi.nlm.nih.gov/pubmed/2476649
- S. Wissing, P. Ludovico, E. Herker, S. Büttner, S.M. Engelhardt, T. Decker, A. Link, A. Proksch, F. Rodrigues, M. Corte-Real, K. Fröhlich, J. Manns, C. Candé, S.J. Sigrist, G. Kroemer, and F. Madeo, "An AIF orthologue regulates apoptosis in yeast", The Journal of Cell Biology, vol. 166, pp. 969-974, 2004. http://dx.doi.org/10.1083/jcb.200404138
- A.L. Hughes, and D.E. Gottschling, "An early age increase in vacuolar pH limits mitochondrial function and lifespan in yeast", Nature, vol. 492, pp. 261-265, 2012. http://dx.doi.org/10.1038/nature11654
- S. Manon, B. Chaudhuri, and M. Guérin, "Release of cytochrome c and decrease of cytochrome c oxidase in Bax-expressing yeast cells, and prevention of these effects by coexpression of Bcl-xL.", FEBS letters, 1997. http://www.ncbi.nlm.nih.gov/pubmed/9326363
- D. Trindade, C. Pereira, S. Chaves, S. Manon, M. Corte-Real, and M. Joao Sousa, "VDAC regulates AAC-mediated apoptosis and cytochrome c release in yeast", Microbial Cell, vol. 3, pp. 500-510, 2016. http://dx.doi.org/10.15698/mic2016.10.533
- C. Pereira, N. Camougrand, S. Manon, M.J. Sousa, and M. Côrte‐Real, "ADP/ATP carrier is required for mitochondrial outer membrane permeabilization and cytochrome c release in yeast apoptosis", Molecular Microbiology, vol. 66, pp. 571-582, 2007. http://dx.doi.org/10.1111/j.1365-2958.2007.05926.x
- S. Giannattasio, A. Atlante, L. Antonacci, N. Guaragnella, P. Lattanzio, S. Passarella, and E. Marra, "Cytochrome c is released from coupled mitochondria of yeast en route to acetic acid‐induced programmed cell death and can work as an electron donor and a ROS scavenger", FEBS Letters, vol. 582, pp. 1519-1525, 2008. http://dx.doi.org/10.1016/j.febslet.2008.03.048
- S. Büttner, D. Carmona-Gutierrez, I. Vitale, M. Castedo, D. Ruli, T. Eisenberg, G. Kroemer, and F. Madeo, "Depletion of endonuclease G selectively kills polyploid cells.", Cell cycle (Georgetown, Tex.), 2007. http://www.ncbi.nlm.nih.gov/pubmed/17471024
- R.E.C. Lee, L.G. Puente, M. Kærn, and L.A. Megeney, "A Non-Death Role of the Yeast Metacaspase: Yca1p Alters Cell Cycle Dynamics", PLoS ONE, vol. 3, pp. e2956, 2008. http://dx.doi.org/10.1371/journal.pone.0002956
- R.E.C. Lee, S. Brunette, L.G. Puente, and L.A. Megeney, "Metacaspase Yca1 is required for clearance of insoluble protein aggregates", Proceedings of the National Academy of Sciences, vol. 107, pp. 13348-13353, 2010. http://dx.doi.org/10.1073/pnas.1006610107
- N. Vahsen, C. Candé, J. Brière, P. Bénit, N. Joza, N. Larochette, P.G. Mastroberardino, M.O. Pequignot, N. Casares, V. Lazar, O. Feraud, N. Debili, S. Wissing, S. Engelhardt, F. Madeo, M. Piacentini, J.M. Penninger, H. Schägger, P. Rustin, and G. Kroemer, "AIF deficiency compromises oxidative phosphorylation", The EMBO Journal, vol. 23, pp. 4679-4689, 2004. http://dx.doi.org/10.1038/sj.emboj.7600461
- S. Büttner, D. Ruli, F. Vögtle, L. Galluzzi, B. Moitzi, T. Eisenberg, O. Kepp, L. Habernig, D. Carmona-Gutierrez, P. Rockenfeller, P. Laun, M. Breitenbach, C. Khoury, K. Fröhlich, G. Rechberger, C. Meisinger, G. Kroemer, and F. Madeo, "A yeast BH3-only protein mediates the mitochondrial pathway of apoptosis", The EMBO Journal, vol. 30, pp. 2779-2792, 2011. http://dx.doi.org/10.1038/emboj.2011.197
- Q. Liang, and B. Zhou, "Copper and Manganese Induce Yeast Apoptosis via Different Pathways", Molecular Biology of the Cell, vol. 18, pp. 4741-4749, 2007. http://dx.doi.org/10.1091/mbc.E07-05-0431
- L. Amigoni, G. Frigerio, E. Martegani, and S. Colombo, "Involvement of Aif1 in apoptosis triggered by lack of Hxk2 in the yeastSaccharomyces cerevisiae", FEMS Yeast Research, vol. 16, pp. fow016, 2016. http://dx.doi.org/10.1093/femsyr/fow016
- N. Guaragnella, A. Bobba, S. Passarella, E. Marra, and S. Giannattasio, "Yeast acetic acid‐induced programmed cell death can occur without cytochrome c release which requires metacaspase YCA1", FEBS Letters, vol. 584, pp. 224-228, 2009. http://dx.doi.org/10.1016/j.febslet.2009.11.072
- D. Carmona-Gutierrez, A. Reisenbichler, P. Heimbucher, M.A. Bauer, R.J. Braun, C. Ruckenstuhl, S. Büttner, T. Eisenberg, P. Rockenfeller, K. Fröhlich, G. Kroemer, and F. Madeo, "Ceramide triggers metacaspase-independent mitochondrial cell death in yeast", Cell Cycle, vol. 10, pp. 3973-3978, 2011. http://dx.doi.org/10.4161/cc.10.22.18212
- V. Longo, M. Ždralević, N. Guaragnella, S. Giannattasio, L. Zolla, and A.M. Timperio, "Proteome and metabolome profiling of wild-type and YCA1 -knock-out yeast cells during acetic acid-induced programmed cell death", Journal of Proteomics, vol. 128, pp. 173-188, 2015. http://dx.doi.org/10.1016/j.jprot.2015.08.003
- A. Rego, M. Costa, S.R. Chaves, N. Matmati, H. Pereira, M.J. Sousa, P. Moradas-Ferreira, Y.A. Hannun, V. Costa, and M. Côrte-Real, "Modulation of Mitochondrial Outer Membrane Permeabilization and Apoptosis by Ceramide Metabolism", PLoS ONE, vol. 7, pp. e48571, 2012. http://dx.doi.org/10.1371/journal.pone.0048571
- P. Hauptmann, and L. Lehle, "Kex1 Protease Is Involved in Yeast Cell Death Induced by Defective N-Glycosylation, Acetic Acid, and Chronological Aging", Journal of Biological Chemistry, vol. 283, pp. 19151-19163, 2008. http://dx.doi.org/10.1074/jbc.M801303200
- D. Wilkinson, and M. Ramsdale, "Proteases and caspase-like activity in the yeast Saccharomyces cerevisiae", Biochemical Society Transactions, vol. 39, pp. 1502-1508, 2011. http://dx.doi.org/10.1042/BST0391502
- H. Yang, Q. Ren, and Z. Zhang, "Cleavage of Mcd1 by Caspase-like Protease Esp1 Promotes Apoptosis in Budding Yeast", Molecular Biology of the Cell, vol. 19, pp. 2127-2134, 2008. http://dx.doi.org/10.1091/mbc.E07-11-1113
- F. Madeo, S. Engelhardt, E. Herker, N. Lehmann, C. Maldener, A. Proksch, S. Wissing, and K. Fröhlich, "Apoptosis in yeast: a new model system with applications in cell biology and medicine", Current Genetics, vol. 41, pp. 208-216, 2002. http://dx.doi.org/10.1007/s00294-002-0310-2
- A. Silva, B. Almeida, B. Sampaio-Marques, M. Reis, S. Ohlmeier, F. Rodrigues, A.D. Vale, and P. Ludovico, "Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is a specific substrate of yeast metacaspase", Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, vol. 1813, pp. 2044-2049, 2011. http://dx.doi.org/10.1016/j.bbamcr.2011.09.010
- P. Mehlen, and D.E. Bredesen, "Dependence Receptors: From Basic Research to Drug Development", Science Signaling, vol. 4, 2011. http://dx.doi.org/10.1126/scisignal.2001521
- Y. Chen, H. Zeng, J. Tian, X. Ban, B. Ma, and Y. Wang, "Dill (Anethum graveolens L.) seed essential oil induces Candida albicans apoptosis in a metacaspase-dependent manner", Fungal Biology, vol. 118, pp. 394-401, 2014. http://dx.doi.org/10.1016/j.funbio.2014.02.004
- F.J. Scariot, L.M. Jahn, J.P. Maianti, A.P.L. Delamare, and S. Echeverrigaray, "The fungicide Mancozeb induces metacaspase-dependent apoptotic cell death in Saccharomyces cerevisiae BY4741", Apoptosis, vol. 21, pp. 866-872, 2016. http://dx.doi.org/10.1007/s10495-016-1251-4
- F. Shirazi, and D. Kontoyiannis, "Micafungin triggers caspase-dependent apoptosis inCandida albicansandCandida parapsilosisbiofilms, including caspofungin non-susceptible isolates", Virulence, vol. 6, pp. 385-394, 2015. http://dx.doi.org/10.1080/21505594.2015.1027479
- X. Wu, W. Chang, A. Cheng, L. Sun, and H. Lou, "Plagiochin E, an antifungal active macrocyclic bis(bibenzyl), induced apoptosis in Candida albicans through a metacaspase-dependent apoptotic pathway", Biochimica et Biophysica Acta (BBA) - General Subjects, vol. 1800, pp. 439-447, 2010. http://dx.doi.org/10.1016/j.bbagen.2010.01.001
- S. Sokolov, A. Pozniakovsky, N. Bocharova, D. Knorre, and F. Severin, "Expression of an expanded polyglutamine domain in yeast causes death with apoptotic markers", Biochimica et Biophysica Acta (BBA) - Bioenergetics, vol. 1757, pp. 660-666, 2006. http://dx.doi.org/10.1016/j.bbabio.2006.05.004
- M.J. Reginato, K.R. Mills, J.K. Paulus, D.K. Lynch, D.C. Sgroi, J. Debnath, S.K. Muthuswamy, and J.S. Brugge, "Integrins and EGFR coordinately regulate the pro-apoptotic protein Bim to prevent anoikis", Nature Cell Biology, vol. 5, pp. 733-740, 2003. http://dx.doi.org/10.1038/ncb1026
- K.J. Verstrepen, and F.M. Klis, "Flocculation, adhesion and biofilm formation in yeasts", Molecular Microbiology, vol. 60, pp. 5-15, 2006. http://dx.doi.org/10.1111/j.1365-2958.2006.05072.x
- A.M. Dranginis, J.M. Rauceo, J.E. Coronado, and P.N. Lipke, "A Biochemical Guide to Yeast Adhesins: Glycoproteins for Social and Antisocial Occasions", Microbiology and Molecular Biology Reviews, vol. 71, pp. 282-294, 2007. http://dx.doi.org/10.1128/MMBR.00037-06
- T. Linder, and C.M. Gustafsson, "Molecular phylogenetics of ascomycotal adhesins—A novel family of putative cell-surface adhesive proteins in fission yeasts", Fungal Genetics and Biology, vol. 45, pp. 485-497, 2008. http://dx.doi.org/10.1016/j.fgb.2007.08.002
- P. Rockenfeller, J. Ring, V. Muschett, A. Beranek, S. Buettner, D. Carmona-Gutierrez, T. Eisenberg, C. Khoury, G. Rechberger, S.D. Kohlwein, G. Kroemer, and F. Madeo, "Fatty acids trigger mitochondrion-dependent necrosis.", Cell cycle (Georgetown, Tex.), 2010. http://www.ncbi.nlm.nih.gov/pubmed/20647757
- J. Santos, M.J. Sousa, and C. Leão, "Ammonium Is Toxic for Aging Yeast Cells, Inducing Death and Shortening of the Chronological Lifespan", PLoS ONE, vol. 7, pp. e37090, 2012. http://dx.doi.org/10.1371/journal.pone.0037090
- P. Scaffidi, T. Misteli, and M.E. Bianchi, "Release of chromatin protein HMGB1 by necrotic cells triggers inflammation", Nature, vol. 418, pp. 191-195, 2002. http://dx.doi.org/10.1038/nature00858
- P. Ludovico, M.J. Sousa, M.T. Silva, C. Leão, and M. Côrte-Real, "Saccharomyces cerevisiae commits to a programmed cell death process in response to acetic acid", Microbiology, vol. 147, pp. 2409-2415, 2001. http://dx.doi.org/10.1099/00221287-147-9-2409
- A.J. Phillips, I. Sudbery, and M. Ramsdale, "Apoptosis induced by environmental stresses and amphotericin B inCandida albicans", Proceedings of the National Academy of Sciences, vol. 100, pp. 14327-14332, 2003. http://dx.doi.org/10.1073/pnas.2332326100
- D. Denton, S. Nicolson, and S. Kumar, "Cell death by autophagy: facts and apparent artefacts", Cell Death & Differentiation, vol. 19, pp. 87-95, 2011. http://dx.doi.org/10.1038/cdd.2011.146
- T.V. Berghe, A. Linkermann, S. Jouan-Lanhouet, H. Walczak, and P. Vandenabeele, "Regulated necrosis: the expanding network of non-apoptotic cell death pathways", Nature Reviews Molecular Cell Biology, vol. 15, pp. 135-147, 2014. http://dx.doi.org/10.1038/nrm3737
- H. Pereira, C. Oliveira, L. Castro, A. Preto, S.R. Chaves, and M. Corte-Real, "Yeast as a tool to explore cathepsin D function", Microbial Cell, vol. 2, pp. 225-234, 2015. http://dx.doi.org/10.15698/mic2015.07.212
- E. Bener Aksam, H. Jungwirth, S.D. Kohlwein, J. Ring, F. Madeo, M. Veenhuis, and I.J. van der Klei, "Absence of the peroxiredoxin Pmp20 causes peroxisomal protein leakage and necrotic cell death", Free Radical Biology and Medicine, vol. 45, pp. 1115-1124, 2008. http://dx.doi.org/10.1016/j.freeradbiomed.2008.07.010
- H. Jungwirth, J. Ring, T. Mayer, A. Schauer, S. Büttner, T. Eisenberg, D. Carmona-Gutierrez, K. Kuchler, and F. Madeo, "Loss of peroxisome function triggers necrosis", FEBS Letters, vol. 582, pp. 2882-2886, 2008. http://dx.doi.org/10.1016/j.febslet.2008.07.023
- H. Kim, A. Kim, and K.W. Cunningham, "Vacuolar H+-ATPase (V-ATPase) Promotes Vacuolar Membrane Permeabilization and Nonapoptotic Death in Stressed Yeast", Journal of Biological Chemistry, vol. 287, pp. 19029-19039, 2012. http://dx.doi.org/10.1074/jbc.M112.363390
- A. Schauer, H. Knauer, C. Ruckenstuhl, H. Fussi, M. Durchschlag, U. Potocnik, and K. Fröhlich, "Vacuolar functions determine the mode of cell death", Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, vol. 1793, pp. 540-545, 2009. http://dx.doi.org/10.1016/j.bbamcr.2008.11.006
- L. Laera, N. Guaragnella, M. Zdralevic, D. Marzulli, Z. Liu, and S. Giannattasio, "The transcription factors ADR1 or CAT8 are required for RTG pathway activation and evasion from yeast acetic acid-induced programmed cell death in raffinose", Microbial Cell, vol. 3, pp. 621-631, 2016. http://dx.doi.org/10.15698/mic2016.12.549
- R.N. Kitsis, and J.D. Molkentin, "Apoptotic cell death “Nixed” by an ER–mitochondrial necrotic pathway", Proceedings of the National Academy of Sciences, vol. 107, pp. 9031-9032, 2010. http://dx.doi.org/10.1073/pnas.1003827107
- A. Vaseva, N. Marchenko, K. Ji, S. Tsirka, S. Holzmann, and U. Moll, "p53 Opens the Mitochondrial Permeability Transition Pore to Trigger Necrosis", Cell, vol. 149, pp. 1536-1548, 2012. http://dx.doi.org/10.1016/j.cell.2012.05.014
- V. Izzo, J.M. Bravo-San Pedro, V. Sica, G. Kroemer, and L. Galluzzi, "Mitochondrial Permeability Transition: New Findings and Persisting Uncertainties", Trends in Cell Biology, vol. 26, pp. 655-667, 2016. http://dx.doi.org/10.1016/j.tcb.2016.04.006
- V.R. Richard, A. Beach, A. Piano, A. Leonov, R. Feldman, M.T. Burstein, P. Kyryakov, A. Gomez-Perez, A. Arlia-Ciommo, S. Baptista, C. Campbell, D. Goncharov, S. Pannu, D. Patrinos, B. Sadri, V. Svistkova, A. Victor, and V.I. Titorenko, "Mechanism of liponecrosis, a distinct mode of programmed cell death", Cell Cycle, vol. 13, pp. 3707-3726, 2014. http://dx.doi.org/10.4161/15384101.2014.965003
- P. Rockenfeller, J. Diessl, and E. al, " Diacylglycerol triggers Rim101 pathway dependent necrosis in yeast: a model for lipotoxicity.", Cell Death Differ in press., 2018.
- A. Silva, B. Sampaio-Marques, �. Fernandes, L. Carreto, F. Rodrigues, M. Holcik, M.A.S. Santos, and P. Ludovico, "Involvement of Yeast HSP90 Isoforms in Response to Stress and Cell Death Induced by Acetic Acid", PLoS ONE, vol. 8, pp. e71294, 2013. http://dx.doi.org/10.1371/journal.pone.0071294
- M. Conrad, J.P.F. Angeli, P. Vandenabeele, and B.R. Stockwell, "Regulated necrosis: disease relevance and therapeutic opportunities", Nature Reviews Drug Discovery, vol. 15, pp. 348-366, 2016. http://dx.doi.org/10.1038/nrd.2015.6
- A. Degterev, Z. Huang, M. Boyce, Y. Li, P. Jagtap, N. Mizushima, G.D. Cuny, T.J. Mitchison, M.A. Moskowitz, and J. Yuan, "Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury", Nature Chemical Biology, vol. 1, pp. 112-119, 2005. http://dx.doi.org/10.1038/nchembio711
- L. Galluzzi, O. Kepp, F.K. Chan, and G. Kroemer, "Necroptosis: Mechanisms and Relevance to Disease", Annual Review of Pathology: Mechanisms of Disease, vol. 12, pp. 103-130, 2017. http://dx.doi.org/10.1146/annurev-pathol-052016-100247
- L. Galluzzi, E.H. Baehrecke, A. Ballabio, P. Boya, J.M. Bravo‐San Pedro, F. Cecconi, A.M. Choi, C.T. Chu, P. Codogno, M.I. Colombo, A.M. Cuervo, J. Debnath, V. Deretic, I. Dikic, E. Eskelinen, G.M. Fimia, S. Fulda, D.A. Gewirtz, D.R. Green, M. Hansen, J.W. Harper, M. Jäättelä, T. Johansen, G. Juhasz, A.C. Kimmelman, C. Kraft, N.T. Ktistakis, S. Kumar, B. Levine, C. Lopez‐Otin, F. Madeo, S. Martens, J. Martinez, A. Melendez, N. Mizushima, C. Münz, L.O. Murphy, J.M. Penninger, M. Piacentini, F. Reggiori, D.C. Rubinsztein, K.M. Ryan, L. Santambrogio, L. Scorrano, A.K. Simon, H. Simon, A. Simonsen, N. Tavernarakis, S.A. Tooze, T. Yoshimori, J. Yuan, Z. Yue, Q. Zhong, and G. Kroemer, "Molecular definitions of autophagy and related processes", The EMBO Journal, vol. 36, pp. 1811-1836, 2017. http://dx.doi.org/10.15252/embj.201796697
- G. Kroemer, and B. Levine, "Autophagic cell death: the story of a misnomer", Nature Reviews Molecular Cell Biology, vol. 9, pp. 1004-1010, 2008. http://dx.doi.org/10.1038/nrm2529
- D. Denton, B. Shravage, R. Simin, K. Mills, D.L. Berry, E.H. Baehrecke, and S. Kumar, "Autophagy, Not Apoptosis, Is Essential for Midgut Cell Death in Drosophila", Current Biology, vol. 19, pp. 1741-1746, 2009. http://dx.doi.org/10.1016/j.cub.2009.08.042
- H. Zhang, and E.H. Baehrecke, "Eaten alive: novel insights into autophagy from multicellular model systems", Trends in Cell Biology, vol. 25, pp. 376-387, 2015. http://dx.doi.org/10.1016/j.tcb.2015.03.001
- G. Das, B.V. Shravage, and E.H. Baehrecke, "Regulation and Function of Autophagy during Cell Survival and Cell Death", Cold Spring Harbor Perspectives in Biology, vol. 4, pp. a008813-a008813, 2012. http://dx.doi.org/10.1101/cshperspect.a008813
- A.L. Anding, and E.H. Baehrecke, "Autophagy in Cell Life and Cell Death", Current Topics in Developmental Biology, pp. 67-91, 2015. http://dx.doi.org/10.1016/bs.ctdb.2015.07.012
- D. Denton, T. Xu, and S. Kumar, "Autophagy as a pro‐death pathway", Immunology & Cell Biology, vol. 93, pp. 35-42, 2014. http://dx.doi.org/10.1038/icb.2014.85
- T. Xu, S. Nicolson, D. Denton, and S. Kumar, "Distinct requirements of Autophagy-related genes in programmed cell death", Cell Death & Differentiation, vol. 22, pp. 1792-1802, 2015. http://dx.doi.org/10.1038/cdd.2015.28
- T. Chang, B.V. Shravage, S.D. Hayes, C.M. Powers, R.T. Simin, J. Wade Harper, and E.H. Baehrecke, "Uba1 functions in Atg7- and Atg3-independent autophagy", Nature Cell Biology, vol. 15, pp. 1067-1078, 2013. http://dx.doi.org/10.1038/ncb2804
- S.A. Dziedzic, and A.B. Caplan, "Identification of autophagy genes participating in zinc-induced necrotic cell death in Saccharomyces cerevisiae.", Autophagy, 2011. http://www.ncbi.nlm.nih.gov/pubmed/21317551
- B. Sampaio-Marques, C. Felgueiras, A. Silva, M. Rodrigues, S. Tenreiro, V. Franssens, A.S. Reichert, T.F. Outeiro, J. Winderickx, and P. Ludovico, "SNCA (α-synuclein)-induced toxicity in yeast cells is dependent on Sir2-mediated mitophagy", Autophagy, vol. 8, pp. 1494-1509, 2012. http://dx.doi.org/10.4161/auto.21275
- M. Leão, S. Gomes, C. Bessa, J. Soares, L. Raimundo, P. Monti, G. Fronza, C. Pereira, and L. Saraiva, "Studying p53 family proteins in yeast: Induction of autophagic cell death and modulation by interactors and small molecules", Experimental Cell Research, vol. 330, pp. 164-177, 2015. http://dx.doi.org/10.1016/j.yexcr.2014.09.028
- G. Serrano-Bueno, A. Hernández, G. López-Lluch, J.R. Pérez-Castiñeira, P. Navas, and A. Serrano, "Inorganic Pyrophosphatase Defects Lead to Cell Cycle Arrest and Autophagic Cell Death through NAD+ Depletion in Fermenting Yeast", Journal of Biological Chemistry, vol. 288, pp. 13082-13092, 2013. http://dx.doi.org/10.1074/jbc.M112.439349
- F. Madeo, A. Zimmermann, M.C. Maiuri, and G. Kroemer, "Essential role for autophagy in life span extension", Journal of Clinical Investigation, vol. 125, pp. 85-93, 2015. http://dx.doi.org/10.1172/JCI73946
- L. Murrow, and J. Debnath, "Autophagy as a Stress-Response and Quality-Control Mechanism: Implications for Cell Injury and Human Disease", Annual Review of Pathology: Mechanisms of Disease, vol. 8, pp. 105-137, 2013. http://dx.doi.org/10.1146/annurev-pathol-020712-163918
- A.L. Alvers, L.K. Fishwick, M.S. Wood, D. Hu, H.S. Chung, W.A. Dunn Jr, and J.P. Aris, "Autophagy and amino acid homeostasis are required for chronological longevity in Saccharomyces cerevisiae", Aging Cell, vol. 8, pp. 353-369, 2009. http://dx.doi.org/10.1111/j.1474-9726.2009.00469.x
- J.P. Aris, A.L. Alvers, R.A. Ferraiuolo, L.K. Fishwick, A. Hanvivatpong, D. Hu, C. Kirlew, M.T. Leonard, K.J. Losin, M. Marraffini, A.Y. Seo, V. Swanberg, J.L. Westcott, M.S. Wood, C. Leeuwenburgh, and W.A. Dunn, "Autophagy and leucine promote chronological longevity and respiration proficiency during calorie restriction in yeast", Experimental Gerontology, vol. 48, pp. 1107-1119, 2013. http://dx.doi.org/10.1016/j.exger.2013.01.006
- P. Boya, R. González-Polo, N. Casares, J. Perfettini, P. Dessen, N. Larochette, D. Métivier, D. Meley, S. Souquere, T. Yoshimori, G. Pierron, P. Codogno, and G. Kroemer, "Inhibition of Macroautophagy Triggers Apoptosis", Molecular and Cellular Biology, vol. 25, pp. 1025-1040, 2005. http://dx.doi.org/10.1128/MCB.25.3.1025-1040.2005
- S. Shen, O. Kepp, M. Michaud, I. Martins, H. Minoux, D. Métivier, M.C. Maiuri, R.T. Kroemer, and G. Kroemer, "Association and dissociation of autophagy, apoptosis and necrosis by systematic chemical study", Oncogene, vol. 30, pp. 4544-4556, 2011. http://dx.doi.org/10.1038/onc.2011.168
- S. Shen, O. Kepp, and G. Kroemer, "The end of autophagic cell death?", Autophagy, vol. 8, pp. 1-3, 2012. http://dx.doi.org/10.4161/auto.8.1.16618
- D. Klionsky, and . et al, " Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition).", 2016.
- R. Torggler, D. Papinski, and C. Kraft, "Assays to Monitor Autophagy in Saccharomyces cerevisiae", Cells, vol. 6, pp. 23, 2017. http://dx.doi.org/10.3390/cells6030023
- K. Kainz, J. Tadic, A. Zimmermann, T. Pendl, D. Carmona-Gutierrez, C. Ruckenstuhl, T. Eisenberg, and F. Madeo, "Methods to Assess Autophagy and Chronological Aging in Yeast", Methods in Enzymology, pp. 367-394, 2017. http://dx.doi.org/10.1016/bs.mie.2016.09.086
- E. Delorme-Axford, R.S. Guimaraes, F. Reggiori, and D.J. Klionsky, "The yeast Saccharomyces cerevisiae: An overview of methods to study autophagy progression", Methods, vol. 75, pp. 3-12, 2015. http://dx.doi.org/10.1016/j.ymeth.2014.12.008
- H. Cheong, T. Yorimitsu, F. Reggiori, J.E. Legakis, C. Wang, and D.J. Klionsky, "Atg17 Regulates the Magnitude of the Autophagic Response", Molecular Biology of the Cell, vol. 16, pp. 3438-3453, 2005. http://dx.doi.org/10.1091/mbc.E04-10-0894
- T. Shintani, and D.J. Klionsky, "Cargo Proteins Facilitate the Formation of Transport Vesicles in the Cytoplasm to Vacuole Targeting Pathway", Journal of Biological Chemistry, vol. 279, pp. 29889-29894, 2004. http://dx.doi.org/10.1074/jbc.M404399200
- H. Cheong, and D.J. Klionsky, "Chapter 1 Biochemical Methods to Monitor Autophagy‐Related Processes in Yeast", Methods in Enzymology, pp. 1-26, 2008. http://dx.doi.org/10.1016/S0076-6879(08)03201-1
- J. Kim, W.P. Huang, and D.J. Klionsky, "Membrane recruitment of Aut7p in the autophagy and cytoplasm to vacuole targeting pathways requires Aut1p, Aut2p, and the autophagy conjugation complex.", The Journal of cell biology, 2001. http://www.ncbi.nlm.nih.gov/pubmed/11149920
- T. Shintani, and F. Reggiori, "Chapter Four Fluorescence Microscopy‐Based Assays for Monitoring Yeast Atg Protein Trafficking", Methods in Enzymology, pp. 43-56, 2008. http://dx.doi.org/10.1016/S0076-6879(08)03204-7
- N. Camougrand, I. Kiššová, B. Salin, and R.J. Devenish, "Chapter 8 Monitoring Mitophagy in Yeast", Methods in Enzymology, pp. 89-107, 2008. http://dx.doi.org/10.1016/S0076-6879(08)03208-4
- C.J. Rosado, D. Mijaljica, I. Hatzinisiriou, M. Prescott, and R.J. Devenish, "Rosella: a fluorescent pH-biosensor for reporting vacuolar turnover of cytosol and organelles in yeast.", Autophagy, 2007. http://www.ncbi.nlm.nih.gov/pubmed/18094608
- S. Subramani, and V. Malhotra, "Non‐autophagic roles of autophagy‐related proteins", EMBO reports, vol. 14, pp. 143-151, 2013. http://dx.doi.org/10.1038/embor.2012.220
- M. Binda, M. Péli-Gulli, G. Bonfils, N. Panchaud, J. Urban, T.W. Sturgill, R. Loewith, and C. De Virgilio, "The Vam6 GEF Controls TORC1 by Activating the EGO Complex", Molecular Cell, vol. 35, pp. 563-573, 2009. http://dx.doi.org/10.1016/j.molcel.2009.06.033
- Y.V. Budovskaya, J.S. Stephan, F. Reggiori, D.J. Klionsky, and P.K. Herman, "The Ras/cAMP-dependent Protein Kinase Signaling Pathway Regulates an Early Step of the Autophagy Process in Saccharomyces cerevisiae", Journal of Biological Chemistry, vol. 279, pp. 20663-20671, 2004. http://dx.doi.org/10.1074/jbc.M400272200
- S. Kira, K. Tabata, K. Shirahama-Noda, A. Nozoe, T. Yoshimori, and T. Noda, "Reciprocal conversion of Gtr1 and Gtr2 nucleotide-binding states by Npr2-Npr3 inactivates TORC1 and induces autophagy", Autophagy, vol. 10, pp. 1565-1578, 2014. http://dx.doi.org/10.4161/auto.29397
- T. Yorimitsu, S. Zaman, J.R. Broach, and D.J. Klionsky, "Protein Kinase A and Sch9 Cooperatively Regulate Induction of Autophagy inSaccharomyces cerevisiae", Molecular Biology of the Cell, vol. 18, pp. 4180-4189, 2007. http://dx.doi.org/10.1091/mbc.E07-05-0485
- Y. Yang, L. Hu, H. Zheng, C. Mao, W. Hu, K. Xiong, F. Wang, and C. Liu, "Application and interpretation of current autophagy inhibitors and activators", Acta Pharmacologica Sinica, vol. 34, pp. 625-635, 2013. http://dx.doi.org/10.1038/aps.2013.5
- J.M. Enserink, H. Hombauer, M. Huang, and R.D. Kolodner, "Cdc28/Cdk1 positively and negatively affects genome stability in S. cerevisiae", Journal of Cell Biology, vol. 185, pp. 423-437, 2009. http://dx.doi.org/10.1083/jcb.200811083
- A. Hibbel, A. Bogdanova, M. Mahamdeh, A. Jannasch, M. Storch, E. Schäffer, D. Liakopoulos, and J. Howard, "Kinesin Kip2 enhances microtubule growth in vitro through length-dependent feedback on polymerization and catastrophe", eLife, vol. 4, 2015. http://dx.doi.org/10.7554/eLife.10542
- M. Kang, S. Yu, H. Lim, B. Choi, C. Park, J. Kang, and S. Lee, "Mitotic catastrophe induced by overexpression of budding yeast Rad2p", Yeast, vol. 27, pp. 399-411, 2010. http://dx.doi.org/10.1002/yea.1764
- D. Lombardi, and L. Lasagni, " Cell-cycle Alterations in Post-mitotic Cells and Cell Death by Mitotic Catastrophe.", In: Najman S, editor Cell Biol. - New Insights. InTech., 2016.
- W.C. Burhans, M. Weinberger, M.A. Marchetti, L. Ramachandran, G. D'Urso, and J.A. Huberman, "Apoptosis-like yeast cell death in response to DNA damage and replication defects.", Mutation research, 2003. http://www.ncbi.nlm.nih.gov/pubmed/14643439
- K. Endo, M. Mizuguchi, A. Harata, G. Itoh, and K. Tanaka, "Nocodazole induces mitotic cell death with apoptotic‐like features in Saccharomyces cerevisiae", FEBS Letters, vol. 584, pp. 2387-2392, 2010. http://dx.doi.org/10.1016/j.febslet.2010.04.029
- M. Weinberger, L. Ramachandran, L. Feng, K. Sharma, X. Sun, M. Marchetti, J.A. Huberman, and W.C. Burhans, "Apoptosis in budding yeast caused by defects in initiation of DNA replication", Journal of Cell Science, vol. 118, pp. 3543-3553, 2005. http://dx.doi.org/10.1242/jcs.02477
- H. Vakifahmetoglu, M. Olsson, and B. Zhivotovsky, "Death through a tragedy: mitotic catastrophe", Cell Death & Differentiation, vol. 15, pp. 1153-1162, 2008. http://dx.doi.org/10.1038/cdd.2008.47
- I. Vitale, L. Galluzzi, M. Castedo, and G. Kroemer, "Mitotic catastrophe: a mechanism for avoiding genomic instability", Nature Reviews Molecular Cell Biology, vol. 12, pp. 385-392, 2011. http://dx.doi.org/10.1038/nrm3115
- P. Fabrizio, and V.D. Longo, "Chronological aging-induced apoptosis in yeast", Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, vol. 1783, pp. 1280-1285, 2008. http://dx.doi.org/10.1016/j.bbamcr.2008.03.017
- C.P. Low, G. Shui, L.P. Liew, S. Buttner, F. Madeo, I.W. Dawes, M.R. Wenk, and H. Yang, "Caspase-dependent and -independent lipotoxic cell-death pathways in fission yeast", Journal of Cell Science, vol. 121, pp. 2671-2684, 2008. http://dx.doi.org/10.1242/jcs.028977
- Q. Zhang, H.K. Chieu, C.P. Low, S. Zhang, C.K. Heng, and H. Yang, "Schizosaccharomyces pombe Cells Deficient in Triacylglycerols Synthesis Undergo Apoptosis upon Entry into the Stationary Phase", Journal of Biological Chemistry, vol. 278, pp. 47145-47155, 2003. http://dx.doi.org/10.1074/jbc.M306998200
- C.P. Low, and H. Yang, "Programmed cell death in fission yeast Schizosaccharomyces pombe", Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, vol. 1783, pp. 1335-1349, 2008. http://dx.doi.org/10.1016/j.bbamcr.2008.02.002
- R. Guérin, G. Arseneault, S. Dumont, and L.A. Rokeach, "Calnexin Is Involved in Apoptosis Induced by Endoplasmic Reticulum Stress in the Fission Yeast", Molecular Biology of the Cell, vol. 19, pp. 4404-4420, 2008. http://dx.doi.org/10.1091/mbc.E08-02-0188
- B. Ink, M. Zörnig, B. Baum, N. Hajibagheri, C. James, T. Chittenden, and G. Evan, "Human Bak induces cell death in Schizosaccharomyces pombe with morphological changes similar to those with apoptosis in mammalian cells.", Molecular and cellular biology, 1997. http://www.ncbi.nlm.nih.gov/pubmed/9111315
- R. Guérin, P.B. Beauregard, A. Leroux, and L.A. Rokeach, "Calnexin Regulates Apoptosis Induced by Inositol Starvation in Fission Yeast", PLoS ONE, vol. 4, pp. e6244, 2009. http://dx.doi.org/10.1371/journal.pone.0006244
- N. MUTOH, S. KITAJIMA, and S. ICHIHARA, "Apoptotic Cell Death in the Fission YeastSchizosaccharomyces pombeInduced by Valproic Acid and Its Extreme Susceptibility to pH Change", Bioscience, Biotechnology, and Biochemistry, vol. 75, pp. 1113-1118, 2011. http://dx.doi.org/10.1271/bbb.110019
- B. Fahrenkrog, U. Sauder, and U. Aebi, "TheS. cerevisiaeHtrA-like protein Nma111p is a nuclear serine protease that mediates yeast apoptosis", Journal of Cell Science, vol. 117, pp. 115-126, 2004. http://dx.doi.org/10.1242/jcs.00848
- C. LOW, L. LIEW, S. PERVAIZ, and H. YANG, "Apoptosis and lipoapoptosis in the fission yeast", FEMS Yeast Research, vol. 5, pp. 1199-1206, 2005. http://dx.doi.org/10.1016/j.femsyr.2005.07.004
- K. Oda, N. Kawasaki, M. Fukuyama, and S. Ikeda, "Ectopic expression of mitochondria endonuclease Pnu1p from Schizosaccharomyces pombe induces cell death of the yeast.", Journal of biochemistry and molecular biology, 2007. http://www.ncbi.nlm.nih.gov/pubmed/18047809
- F.C. Odds, A.J. Brown, and N.A. Gow, "Array", Genome Biology, vol. 5, pp. 230, 2004. http://dx.doi.org/10.1186/gb-2004-5-7-230
- J. Kim, and P. Sudbery, "Candida albicans, a major human fungal pathogen", The Journal of Microbiology, vol. 49, pp. 171-177, 2011. http://dx.doi.org/10.1007/s12275-011-1064-7
- K. De Brucker, B.P. Cammue, and K. Thevissen, "Apoptosis-inducing antifungal peptides and proteins", Biochemical Society Transactions, vol. 39, pp. 1527-1532, 2011. http://dx.doi.org/10.1042/BST0391527
- S. Lin, and N. Austriaco, "Aging and cell death in the other yeasts,Schizosaccharomyces pombeandCandida albicans", FEMS Yeast Research, vol. 14, pp. 119-135, 2013. http://dx.doi.org/10.1111/1567-1364.12113
- M. Ramsdale, "Programmed cell death in pathogenic fungi", Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, vol. 1783, pp. 1369-1380, 2008. http://dx.doi.org/10.1016/j.bbamcr.2008.01.021
- M.S. Ahmadi, H.H. Lee, D.A. Sanchez, A.J. Friedman, M.T. Tar, K.P. Davies, J.D. Nosanchuk, and L.R. Martinez, "Sustained Nitric Oxide-Releasing Nanoparticles Induce Cell Death in Candida albicans Yeast and Hyphal Cells, Preventing Biofilm Formation In Vitro and in a Rodent Central Venous Catheter Model", Antimicrobial Agents and Chemotherapy, vol. 60, pp. 2185-2194, 2016. http://dx.doi.org/10.1128/AAC.02659-15
- R.S. Al-Dhaheri, and L.J. Douglas, "Apoptosis in Candida biofilms exposed to amphotericin B", Journal of Medical Microbiology, vol. 59, pp. 149-157, 2010. http://dx.doi.org/10.1099/jmm.0.015784-0
- V.S. Thibane, R. Ells, A. Hugo, J. Albertyn, W.J.J. van Rensburg, P.W. Van Wyk, J.L. Kock, and C.H. Pohl, "Polyunsaturated fatty acids cause apoptosis in C. albicans and C. dubliniensis biofilms", Biochimica et Biophysica Acta (BBA) - General Subjects, vol. 1820, pp. 1463-1468, 2012. http://dx.doi.org/10.1016/j.bbagen.2012.05.004
- Y. Cao, S. Huang, B. Dai, Z. Zhu, H. Lu, L. Dong, Y. Cao, Y. Wang, P. Gao, Y. Chai, and Y. Jiang, "Candida albicans cells lacking CaMCA1-encoded metacaspase show resistance to oxidative stress-induced death and change in energy metabolism", Fungal Genetics and Biology, vol. 46, pp. 183-189, 2009. http://dx.doi.org/10.1016/j.fgb.2008.11.001
- M.E. Shirtliff, B.P. Krom, R.A.M. Meijering, B.M. Peters, J. Zhu, M.A. Scheper, M.L. Harris, and M.A. Jabra-Rizk, "Farnesol-Induced Apoptosis in Candida albicans", Antimicrobial Agents and Chemotherapy, vol. 53, pp. 2392-2401, 2009. http://dx.doi.org/10.1128/AAC.01551-08
- B. Hao, S. Cheng, C.J. Clancy, and M.H. Nguyen, "Caspofungin Kills Candida albicans by Causing both Cellular Apoptosis and Necrosis", Antimicrobial Agents and Chemotherapy, vol. 57, pp. 326-332, 2013. http://dx.doi.org/10.1128/AAC.01366-12
- V. Cabezón, V. Vialás, A. Gil-Bona, J.A. Reales-Calderón, M. Martínez-Gomariz, D. Gutiérrez-Blázquez, L. Monteoliva, G. Molero, M. Ramsdale, and C. Gil, "Apoptosis of Candida albicans during the Interaction with Murine Macrophages: Proteomics and Cell-Death Marker Monitoring", Journal of Proteome Research, vol. 15, pp. 1418-1434, 2016. http://dx.doi.org/10.1021/acs.jproteome.5b00913
- A.M. Aerts, D. Carmona-Gutierrez, S. Lefevre, G. Govaert, I.E. François, F. Madeo, R. Santos, B.P. Cammue, and K. Thevissen, "The antifungal plant defensin RsAFP2 from radish induces apoptosis in a metacaspase independent way in Candida albicans", FEBS Letters, vol. 583, pp. 2513-2516, 2009. http://dx.doi.org/10.1016/j.febslet.2009.07.004
- A.J. Phillips, J.D. Crowe, and M. Ramsdale, "Ras pathway signaling accelerates programmed cell death in the pathogenic fungus Candida albicans", Proceedings of the National Academy of Sciences, vol. 103, pp. 726-731, 2006. http://dx.doi.org/10.1073/pnas.0506405103
- B. Dai, Y. Cao, S. Huang, Y. Xu, P. Gao, Y. Wang, and Y. Jiang, "Baicalein induces programmed cell death in Candida albicans.", Journal of microbiology and biotechnology, 2009. http://www.ncbi.nlm.nih.gov/pubmed/19734718
-
B. Dai, Y. Wang, L. Zhao, D. Li, M. Li, Y. Cao, and Y. Jiang, "Cap1p attenuates the apoptosis of
C andida albicans", The FEBS Journal, vol. 280, pp. 2633-2643, 2013. http://dx.doi.org/10.1111/febs.12251 - H. Almshawit, D. Pouniotis, and I. Macreadie, "Cell density impacts onCandida glabratasurvival in hypo-osmotic stress", FEMS Yeast Research, vol. 14, pp. 508-516, 2013. http://dx.doi.org/10.1111/1567-1364.12122
- K. Kang, K. Wong, W. Fong, and P.W. Tsang, "Metergoline-induced cell death in Candida krusei", Fungal Biology, vol. 115, pp. 302-309, 2011. http://dx.doi.org/10.1016/j.funbio.2011.01.001
- P.W. Tsang, A.P. Wong, H. Yang, and N. Li, "Purpurin Triggers Caspase-Independent Apoptosis in Candida dubliniensis Biofilms", PLoS ONE, vol. 8, pp. e86032, 2013. http://dx.doi.org/10.1371/journal.pone.0086032
- C.R. da Silva, J.B. de Andrade Neto, R. de Sousa Campos, N.S. Figueiredo, L.S. Sampaio, H.I.F. Magalhães, B.C. Cavalcanti, D.M. Gaspar, G.M. de Andrade, I.S.P. Lima, G.S. de Barros Viana, M.O. de Moraes, M.D.P. Lobo, T.B. Grangeiro, and H.V. Nobre Júnior, "Synergistic Effect of the Flavonoid Catechin, Quercetin, or Epigallocatechin Gallate with Fluconazole Induces Apoptosis in Candida tropicalis Resistant to Fluconazole", Antimicrobial Agents and Chemotherapy, vol. 58, pp. 1468-1478, 2014. http://dx.doi.org/10.1128/AAC.00651-13
- F. Shirazi, R. Lewis, and D. Kontoyiannis, "Micafungin induced apoptosis in Candida parapsilosis independent of its susceptibility to micafungin", Microbial Cell, vol. 2, pp. 445-450, 2015. http://dx.doi.org/10.15698/mic2015.11.236
- R. Ikeda, and K. Sawamura, "Bacterial and H2O2 stress-induced apoptosis-like events in Cryptococcus neoformans", Research in Microbiology, vol. 159, pp. 628-634, 2008. http://dx.doi.org/10.1016/j.resmic.2008.07.006
- C.P. Semighini, A.F. Averette, J.R. Perfect, and J. Heitman, "Deletion of Cryptococcus neoformans AIF Ortholog Promotes Chromosome Aneuploidy and Fluconazole-Resistance in a Metacaspase-Independent Manner", PLoS Pathogens, vol. 7, pp. e1002364, 2011. http://dx.doi.org/10.1371/journal.ppat.1002364
- Y. Wang, and Z. Wang, "Sodium citrate induces apoptosis in biocontrol yeast Cryptococcus laurentii", Journal of Applied Microbiology, vol. 113, pp. 135-142, 2012. http://dx.doi.org/10.1111/j.1365-2672.2012.05312.x
- C. Mazzoni, P. Mancini, F. Madeo, V. Palermo, and C. Falcone, "A Kluyveromyces lactis mutant in the essential gene KlLSM4 shows phenotypic markers of apoptosis.", FEMS yeast research, 2003. http://www.ncbi.nlm.nih.gov/pubmed/14554194
- D. Poliaková, B. Sokolı́ková, J. Kolarov, and L. Šabova, "The antiapoptotic protein Bcl-xL prevents the cytotoxic effect of Bax, but not Bax-induced formation of reactive oxygen species, in Kluyveromyces lactis", Microbiology, vol. 148, pp. 2789-2795, 2002. http://dx.doi.org/10.1099/00221287-148-9-2789
- S. Abdelmoula-Souissi, I. Mabrouk, A. Gargouri, and R. Mokdad-Gargouri, "Expression of the human tumor suppressor p53 induces cell death in Pichia pastoris", FEMS Yeast Research, vol. 12, pp. 2-8, 2011. http://dx.doi.org/10.1111/j.1567-1364.2011.00758.x
- J. Chen, B. Li, G. Qin, and S. Tian, "Mechanism of H2O2-induced oxidative stress regulating viability and biocontrol ability of Rhodotorula glutinis", International Journal of Food Microbiology, vol. 193, pp. 152-158, 2015. http://dx.doi.org/10.1016/j.ijfoodmicro.2014.10.025
- P. Ludovico, F. Sansonetty, M.T. Silva, and M. Côrte-Real, "Acetic acid induces a programmed cell death process in the food spoilage yeast Zygosaccharomyces bailii.", FEMS yeast research, 2003. http://www.ncbi.nlm.nih.gov/pubmed/12702251
- J. Guerreiro, B. Sampaio-Marques, R. Soares, A. Coelho, C. Leao, P. Ludovico, and I. Sa-Correia, "Mitochondrial proteomics of the acetic acid - induced programmed cell death response in a highly tolerant Zygosaccharomyces bailii - derived hybrid strain", Microbial Cell, vol. 3, pp. 65-78, 2016. http://dx.doi.org/10.15698/mic2016.02.477
- D. Carmona-Gutierrez, C. Sommer, A. Andryushkova, G. Kroemer, and F. Madeo, "A higher spirit: avoiding yeast suicide during alcoholic fermentation", Cell Death & Differentiation, vol. 19, pp. 913-914, 2012. http://dx.doi.org/10.1038/cdd.2012.31
- K.Z. Hein, H. Takahashi, T. Tsumori, Y. Yasui, Y. Nanjoh, T. Toga, Z. Wu, J. Grötzinger, S. Jung, J. Wehkamp, B.O. Schroeder, J.M. Schroeder, and E. Morita, "Disulphide-reduced psoriasin is a human apoptosis-inducing broad-spectrum fungicide", Proceedings of the National Academy of Sciences, vol. 112, pp. 13039-13044, 2015. http://dx.doi.org/10.1073/pnas.1511197112
- S.A.A. Mousavi, and G.D. Robson, "Entry into the stationary phase is associated with a rapid loss of viability and an apoptotic-like phenotype in the opportunistic pathogen Aspergillus fumigatus.", Fungal genetics and biology : FG & B, 2003. http://www.ncbi.nlm.nih.gov/pubmed/12892635
- S.A.A. Mousavi, and G.D. Robson, "Oxidative and amphotericin B-mediated cell death in the opportunistic pathogen Aspergillus fumigatus is associated with an apoptotic-like phenotype", Microbiology, vol. 150, pp. 1937-1945, 2004. http://dx.doi.org/10.1099/mic.0.26830-0
-
K. Fujita, M. Tatsumi, A. Ogita, I. Kubo, and T. Tanaka, "Anethole induces apoptotic cell death accompanied by reactive oxygen species production and
DNA fragmentation inA spergillus fumigatus andS accharomyces cerevisiae", The FEBS Journal, vol. 281, pp. 1304-1313, 2014. http://dx.doi.org/10.1111/febs.12706 - D.L. Richie, M.D. Miley, R. Bhabhra, G.D. Robson, J.C. Rhodes, and D.S. Askew, "The Aspergillus fumigatus metacaspases CasA and CasB facilitate growth under conditions of endoplasmic reticulum stress", Molecular Microbiology, vol. 63, pp. 591-604, 2006. http://dx.doi.org/10.1111/j.1365-2958.2006.05534.x
- J. Cheng, T. Park, L. Chio, A.S. Fischl, and X.S. Ye, "Induction of apoptosis by sphingoid long-chain bases in Aspergillus nidulans.", Molecular and cellular biology, 2003. http://www.ncbi.nlm.nih.gov/pubmed/12482970
- M. Savoldi, I. Malavazi, F.M. Soriani, J.L. Capellaro, K. Kitamoto, M.E. da Silva Ferreira, M.H.S. Goldman, and G.H. Goldman, "Farnesol induces the transcriptional accumulation of the Aspergillus nidulans Apoptosis‐Inducing Factor (AIF)‐like mitochondrial oxidoreductase", Molecular Microbiology, vol. 70, pp. 44-59, 2008. http://dx.doi.org/10.1111/j.1365-2958.2008.06385.x
- C. Thrane, U. Kaufmann, B. Stummann, and S. Olsson, "Activation of caspase-like activity and poly (ADP-ribose) polymerase degradation during sporulation in Aspergillus nidulans", Fungal Genetics and Biology, vol. 41, pp. 361-368, 2004. http://dx.doi.org/10.1016/j.fgb.2003.11.003
- D. Brust, A. Hamann, and H.D. Osiewacz, "Deletion of PaAif2 and PaAmid2, two genes encoding mitochondrial AIF-like oxidoreductases of Podospora anserina, leads to increased stress tolerance and lifespan extension", Current Genetics, vol. 56, pp. 225-235, 2010. http://dx.doi.org/10.1007/s00294-010-0295-1
- A. Hamann, D. Brust, and H.D. Osiewacz, "Deletion of putative apoptosis factors leads to lifespan extension in the fungal ageing model Podospora anserina", Molecular Microbiology, vol. 65, pp. 948-958, 2007. http://dx.doi.org/10.1111/j.1365-2958.2007.05839.x
- D. Brust, B. Daum, C. Breunig, A. Hamann, W. Kühlbrandt, and H.D. Osiewacz, "Cyclophilin D links programmed cell death and organismal aging in Podospora anserina", Aging Cell, vol. 9, pp. 761-775, 2010. http://dx.doi.org/10.1111/j.1474-9726.2010.00609.x
- B. Daum, A. Walter, A. Horst, H.D. Osiewacz, and W. Kühlbrandt, "Age-dependent dissociation of ATP synthase dimers and loss of inner-membrane cristae in mitochondria", Proceedings of the National Academy of Sciences, vol. 110, pp. 15301-15306, 2013. http://dx.doi.org/10.1073/pnas.1305462110
- P. Kramer, A.T. Jung, A. Hamann, and H.D. Osiewacz, "Cyclophilin D Is Involved in the Regulation of Autophagy and Affects the Lifespan of P. anserina in Response to Mitochondrial Oxidative Stress", Frontiers in Genetics, vol. 7, 2016. http://dx.doi.org/10.3389/fgene.2016.00165
- L. Knuppertz, A. Hamann, F. Pampaloni, E. Stelzer, and H.D. Osiewacz, "Identification of autophagy as a longevity-assurance mechanism in the aging modelPodospora anserina", Autophagy, vol. 10, pp. 822-834, 2014. http://dx.doi.org/10.4161/auto.28148
-
L. Knuppertz, and H.D. Osiewacz, "Autophagy compensates impaired energy metabolism in
CLPXP ‐deficient Podospora anserina strains and extends healthspan", Aging Cell, vol. 16, pp. 704-715, 2017. http://dx.doi.org/10.1111/acel.12600 - A. Sharon, A. Finkelstein, N. Shlezinger, and I. Hatam, "Fungal apoptosis: function, genes and gene function", FEMS Microbiology Reviews, vol. 33, pp. 833-854, 2009. http://dx.doi.org/10.1111/j.1574-6976.2009.00180.x
- D.C. Arruda, A.L. Matsuo, L.S. Silva, F. Real, N.P. Leitão, J.H.S. Pires, A.C.F. Caires, D.M. Garcia, F.F.M. Cunha, R. Puccia, and L.V.G. Longo, "Cyclopalladated Compound 7a Induces Apoptosis- and Autophagy-Like Mechanisms in Paracoccidioides and Is a Candidate for Paracoccidioidomycosis Treatment", Antimicrobial Agents and Chemotherapy, vol. 59, pp. 7214-7223, 2015. http://dx.doi.org/10.1128/AAC.00512-15
- S. Barhoom, and A. Sharon, "Bcl-2 proteins link programmed cell death with growth and morphogenetic adaptations in the fungal plant pathogen Colletotrichum gloeosporioides", Fungal Genetics and Biology, vol. 44, pp. 32-43, 2007. http://dx.doi.org/10.1016/j.fgb.2006.06.007
- S. Ito, T. Ihara, H. Tamura, S. Tanaka, T. Ikeda, H. Kajihara, C. Dissanayake, F.F. Abdel-Motaal, and M.A. El-Sayed, "α‐Tomatine, the major saponin in tomato, induces programmed cell death mediated by reactive oxygen species in the fungal pathogen Fusarium oxysporum", FEBS Letters, vol. 581, pp. 3217-3222, 2007. http://dx.doi.org/10.1016/j.febslet.2007.06.010
- C.P. Semighini, N. Murray, and S.D. Harris, "Inhibition ofFusarium graminearumgrowth and development by farnesol", FEMS Microbiology Letters, vol. 279, pp. 259-264, 2008. http://dx.doi.org/10.1111/j.1574-6968.2007.01042.x
- L. Roze, and J. Linz, "Lovastatin Triggers an Apoptosis-like Cell Death Process in the FungusMucor racemosus", Fungal Genetics and Biology, vol. 25, pp. 119-133, 1998. http://dx.doi.org/10.1006/fgbi.1998.1093
- A. Finkelshtein, N. Shlezinger, O. Bunis, and A. Sharon, "Botrytis cinerea BcNma is involved in apoptotic cell death but not in stress adaptation", Fungal Genetics and Biology, vol. 48, pp. 621-630, 2011. http://dx.doi.org/10.1016/j.fgb.2011.01.007
- P. Liu, L. Luo, J. Guo, H. Liu, B. Wang, B. Deng, C. Long, and Y. Cheng, "Farnesol induces apoptosis and oxidative stress in the fungal pathogenPenicillium expansum", Mycologia, vol. 102, pp. 311-318, 2010. http://dx.doi.org/10.3852/09-176
- F. Shirazi, M.A. Pontikos, T.J. Walsh, N. Albert, R.E. Lewis, and D.P. Kontoyiannis, "Hyperthermia Sensitizes Rhizopus oryzae to Posaconazole and Itraconazole Action through Apoptosis", Antimicrobial Agents and Chemotherapy, vol. 57, pp. 4360-4368, 2013. http://dx.doi.org/10.1128/AAC.00571-13
- F. Shirazi, . , and D. Kontoyiannis, "Heat shock protein 90 and calcineurin pathway inhibitors enhance the efficacy of triazoles against Scedosporium prolificans via induction of apoptosis", Microbial Cell, vol. 1, pp. 179-188, 2014. http://dx.doi.org/10.15698/mic2014.06.150
- P. Carneiro, M. Duarte, and A. Videira, "Characterization of Apoptosis-Related Oxidoreductases from Neurospora crassa", PLoS ONE, vol. 7, pp. e34270, 2012. http://dx.doi.org/10.1371/journal.pone.0034270
- N.D. Fedorova, J.H. Badger, G.D. Robson, J.R. Wortman, and W.C. Nierman, "Comparative analysis of programmed cell death pathways in filamentous fungi", BMC Genomics, vol. 6, 2005. http://dx.doi.org/10.1186/1471-2164-6-177
AUTHOR CONTRIBUTIONS
DCG, MAB, LG, GK and FM conceived and wrote the manu-script. All authors participated in the critical reading of the manuscript, completed or approved the cited literature, ap-proved the conclusions, and endorse the consensus of the herein defined terminology.
COPYRIGHT
© 2018

Guidelines and recommendations on yeast cell death nomenclature by Carmona-Gutierrez et al. is licensed under a Creative Commons Attribution 4.0 International License.