Back to article: Molecular signature of the imprintosome complex at the mating-type locus in fission yeast
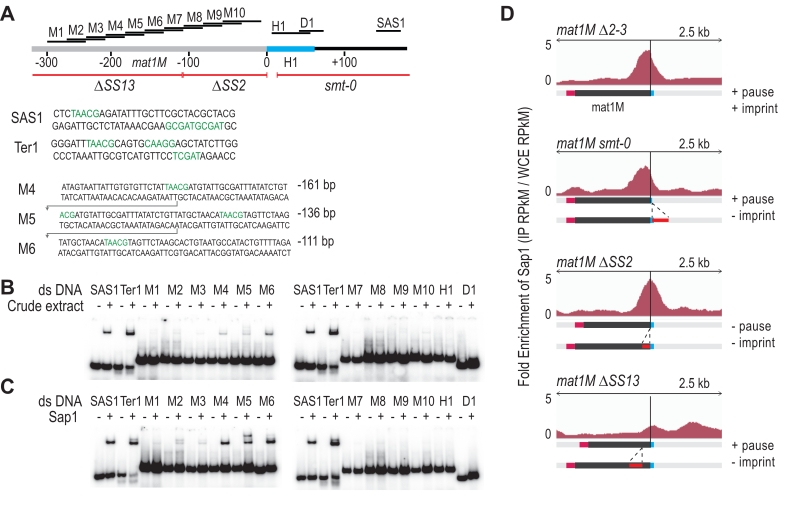
FIGURE 5: Sap1 is recruited in mat1M. (A) Schematic view of right region of the mat1M locus. The probes used for the shift assay as well as the sites of the deletion strains used for the ChIP-seq of Sap1 are indicated. The sequence of the probes that trigger a shift in B and C are indicated. (B) Electrophoretic mobility shift analysis (EMSA) of crude extracts from S. pombe. SAS1 and Ter1 are used as controls. In lanes (-), 1 ng of radio labelled dsDNA is used with no proteins. In lanes (+), 5 µg of total proteins and 1 ng of radio labelled ds DNA are used. (C) EMSA of Sap1-6xHis protein purified from E. coli. In lanes (-), 1 ng of radio labelled ds DNA is used with no proteins. In lanes (+), 40 ng of Sap1-6xHis protein and 1 ng of radio labelled ds DNA are used. (D) Distribution of normalized enrichments of Sap1 ChIPs (IP RPkM / WCE RPkM) in the mat1M ∆2-3, mat1M smt-0 ∆2-3, mat1M ∆SS2 ∆2-3 and mat1M ∆SS13 ∆2-3 backgrounds. The sequence used for the alignment is a 10-kb region that contains the mat1M, mat1M smt-0, mat1M ∆SS2 or mat1M ∆SS13, respectively. Only mat1M is shown. The MT switching phenotype of the deletion strains is indicated.