Back to article: The transcription factors ADR1 or CAT8 are required for RTG pathway activation and evasion from yeast acetic acid-induced programmed cell death in raffinose
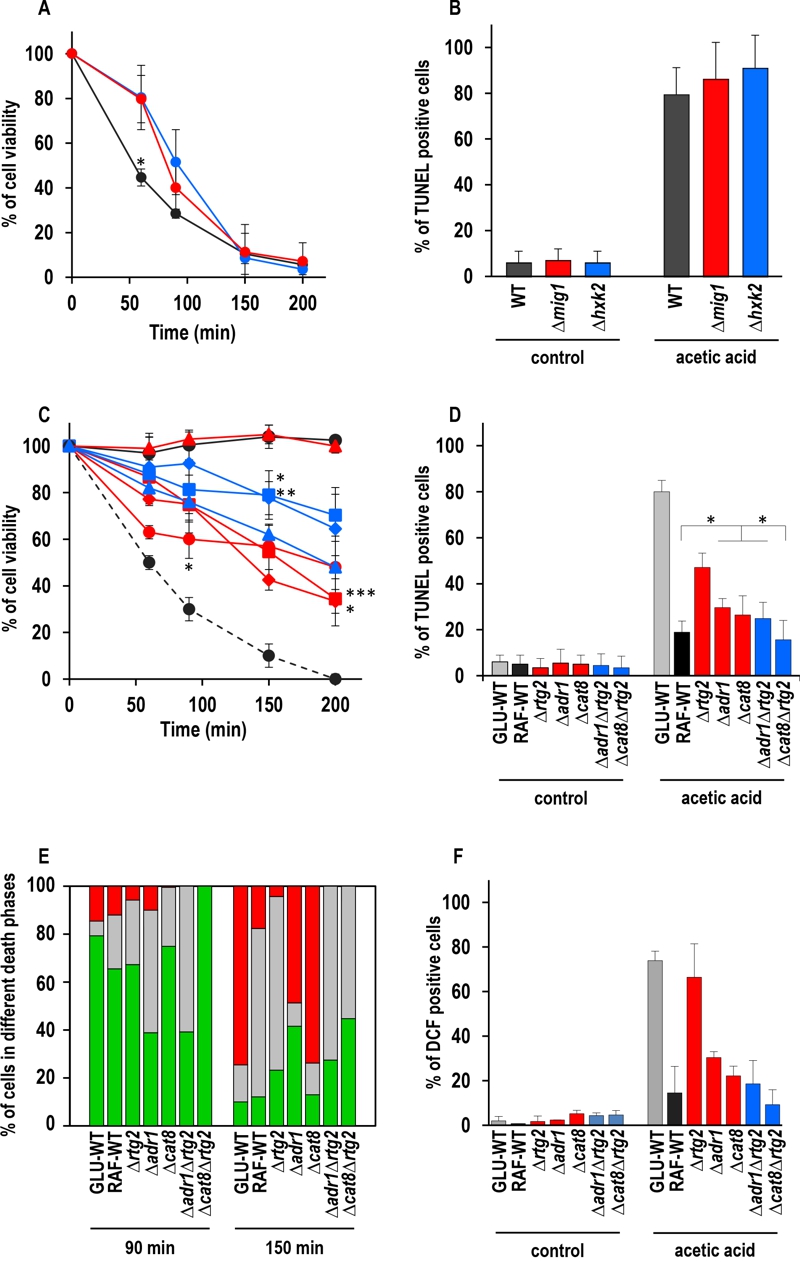
FIGURE 2: Effect of genetic inactivation of both RTG and CCR pathways on AA-PCD in either glucose- or raffinose-grown cells.
(A) Wild-type (WT, black), Δmig1 (red) and Δhxk2 (blue) mutant cells were treated with 80 mM acetic acid in growth medium with glucose as carbon source. Cell viability was analyzed by measuring colony-forming units (cfu) at indicated times. Cell survival based on the cfu was set at 100% at 0 min. The means of five independent experiments with standard deviations are reported. Anova-Bonferroni test: statistically different with (*) p < 0.001 when comparing WT with Δmig1 or Δhxk2 mutant cells.
(B) DNA fragmentation in cells grown in glucose was detected by the TUNEL assay using confocal microscopy analysis. Percentage of TUNEL-positive cells is reported at 150 min. At least 400 cells were analyzed in three samples from each of three independent experiments. WT, grey bars; Δmig1, red bars; Δhxk2, blue bars.
(C) GLU-WT (●, dashed line) and RAF-WT (●, black line), Δrtg2 (●, red line), Δadr1 (■, red line), Δcat8 (♦, red line), Δhap4 (▲, red line), Δadr1Δrtg2 (■, blue line), Δcat8Δrtg2 (♦, blue line) and Δhap4Δrtg2 (▲, blue line) cells were treated with acetic acid either in glucose or in raffinose as carbon source. Cell viability was analyzed at indicated times by measuring colony-forming units (cfu). Cell survival (100%) corresponds to the cfu at time zero. The means of five independent experiments with standard deviations are reported. Anova-Bonferroni test: statistically different with (*) p < 0.01 when comparing Δrtg2 versus Δcat8Δrtg2 at 90 min, Δadr1 with Δadr1Δrtg2 at 150 min or both Δadr1 and Δcat8 with Δcat8Δrtg2 at 200 min; (**) p < 0.001 when comparing Δcat8 with Δcat8Δrtg2 at 150 min; (***) p < 0.0001 when comparing Δadr1 with Δadr1Δrtg2 at 200 min.
(D) DNA fragmentation was detected by TUNEL assay using confocal microscopy analysis. Percentage of TUNEL-positive cells is reported at 150 min. At least 400 cells were analyzed in three samples from each of three independent experiments. Fisher’s exact test: statistically different with (*) p < 0.01 when comparing Δadr1, Δcat8 or Δadr1Δrtg2 versus WT or Δcat8. GLU-WT, grey bars; RAF-WT, black bars; single knock-out cells, red bars; double knock-out cells, blue bars.
(E) In a typical experiment GLU-WT, RAF-WT and knock-out cells grown in raffinose as indicated, were treated with acetic acid (AA). The cells were collected at 90 and 150 min after AA treatment, co-stained with Annexin V-FITC/PI and analyzed by confocal microscopy to measure PS externalization on the cell surface. The bars indicate the percentage of stained cells at different stages of death: early apoptotic cells (Annexin V+/PI–, green); late apoptotic cells (Annexin V+/PI+, grey); necrotic cells (Annexin V–/PI+, red). At least 400 cells were counted for each sample at indicated time. The experiment was repeated twice, with virtual identical results. Fisher’s exact test: statistically different with p < 0.005 when comparing Annexin V+ Δadr1 or Δadr1Δrtg2 versus all other cell types at 90 min, when comparing PI+ Δadr1 versus Δcat8 at 150 min and when comparing Annexin V+ Δadr1 or Δcat8Δrtg2 versus all other cell types at 150 min.
(F) GLU-WT, RAF-WT and knock-out cells grown in raffinose, as indicated, were incubated in the absence (control) or in the presence of acetic acid for 30 min, then collected and stained with H2DCF-DA. DCF-stained cells due to ROS accumulation were analyzed by confocal microscopy. Bars indicate the percentage of DCF-positive cells with SD, calculated by counting at least 400 cells in three independent experiments. GLU-WT, grey bars; RAF-WT, black bars; single knock-out cells, red bars; double knock-out cells, blue bars.