Back to article: Basal level of ppGpp coordinates Escherichia coli cell heterogeneity and ampicillin resistance and persistence
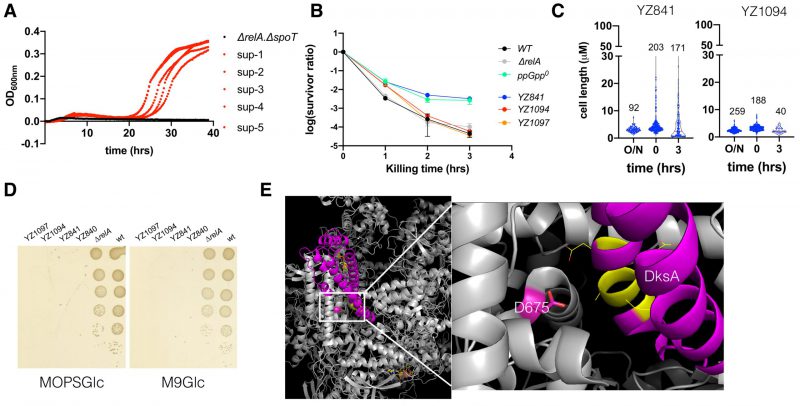
FIGURE 6: The ppGpp0 suppressing mutants rescued the ampicillin MIC and are less heterogeneous and persistent. (A) Growth curves of the parental ΔrelA ΔspoT strain and the five suppressor strains in the presence of 12.5 μg/ml ampicillin. (B) The ampicillin (250 μg/ml) killing curves of the parental ΔrelA ΔspoT strain (YZ841) and the two suppressor strains (YZ1094, YZ1097). Three biological replicates were performed, and the average and SEM were presented. For comparison, the same killing curves of the three strains from (Figure 2B) were included. (C) Box plot of the cell length (µm) of the parental ΔrelA ΔspoT strain and the suppressor strain YZ1094 cells during regrowth and ampicillin killing. The numbers of analyzed cells are indicated above the plots. (D) Growth of the parental ΔrelA ΔspoT(YZ840/YZ841) and the suppressor strains (YZ1094/YZ1097) on MOPS (left) and M9 (right) minimal media agar plate supplemented with glucose 0.2% g/ml (MOPSGlc and M9Glc). Pictures were taken after 24 hrs growth at 37°C. (E) 3D position of the suppressing mutation D675A on RpoB, in a complex structure of E. coli RNAP (grey), DksA (magenta), and two molecules of ppGpp (yellow stick model) (PDB 5VSW) [50]). D675 is shown in a stick model (magenta). To the right side is a zoom-in of the framed region, with DksA residues directly facing D675 shown in yellow and line model.
50. Molodtsov V, Sineva E, Zhang L, Huang X, Cashel M, Ades SE, Murakami KS (2018). Allosteric Effector ppGpp Potentiates the Inhibition of Transcript Initiation by DksA. Mol Cell 69(5): 828-839 e825. 10.1016/j.molcel.2018.01.035