Research Reports:
Microbial Cell, Vol. 11, No. 1, pp. 378 - 386; doi: 10.15698/mic2024.11.840
Microwave-assisted preparation of yeast cells for ultrastructural analysis by electron microscopy
Zellbiologie und Elektronenmikroskopie, Universität Bayreuth, 95440 Bayreuth, Germany.
a These authors contributed equally to this work.
Keywords: microwave fixation, mitochondria, organelle architecture, potassium permanganate fixation, transmission electron microscopy, Saccharomyces cerevisiae.
Received originally: 14/08/2024 Received in revised form: 21/10/2024
Accepted: 24/10/2024
Published: 18/11/2024
Correspondence:
Till Klecker, Zellbiologie und Elektronenmikroskopie, Universität Bayreuth, 95440 Bayreuth, Germany; Phone: +49-921 55 4311; till.klecker@uni-bayreuth.de
Benedikt Westermann, Zellbiologie und Elektronenmikroskopie, Universität Bayreuth, 95440 Bayreuth, Germany; Phone: +49-921 55 4300; benedikt.westermann@uni-bayreuth.de
Conflict of interest statement: The authors declare that there are no conflicts of interest.
Please cite this article as: Moritz Mayer, Christina Schug, Stefan Geimer, Till Klecker and Benedikt Westermann (2024). Microwave-assisted preparation of yeast cells for ultrastructural analysis by electron microscopy. Microbial Cell 11: 378-386. doi: 10.15698/mic2024.11.840
Abstract
Budding yeast Saccharomyces cerevisiae is widely used as a model organism to study the biogenesis and architecture of organellar membranes, which can be visualized by transmission electron microscopy (TEM). Preparation of yeast cells for TEM can be quite challenging and time-consuming. Here, we describe an optimized protocol for conventional fixation of yeast cells with potassium permanganate combined with cell wall permeabilization with sodium metaperiodate and embedding in Epon. We have replaced time-consuming incubation steps by short treatments with microwaves and developed a microwave-assisted permanganate fixation and Epon embedding protocol that reduces the time required for sample preparation to one working day. We expect that these protocols will be useful for routine analysis of membrane ultrastructure in yeast.
INTRODUCTION
Budding yeast Saccharomyces cerevisiae is an excellent model organism for the study of organelle biogenesis in eukaryotic cells1, 2. Seminal discoveries have been made in yeast, including the discovery of genes that control the secretory pathway 3, 4 or encode the machinery of autophagy 5, 6. More recently, the evolutionarily conserved machinery that shapes the mitochondrial inner membrane has been described in yeast 7, 8, 9, 10. These and many other important breakthroughs relied on the visualization of cellular membranes by transmission electron microscopy (TEM).
The analysis of yeast ultrastructure by TEM can be quite challenging. The cell wall impairs infiltration of the cell with chemicals and resins, and the protein concentration in the cytosol is high, which often results in poor contrast and insufficient morphological resolution 11, 12. A growing number of different methods for sample preparation have been developed, including chemical and cryo methods for fixation and a variety of different approaches for embedding and membrane contrasting (reviewed in 11, 13, 14). The choice of the appropriate method depends primarily on the scientific question, i.e. whether further processing, such as immunolabelling, is required, and on the resolution and contrast needed to reliably visualize the cellular structures of interest. Other important factors are the availability of specific instruments and equipment, possible time constraints and also personal skills and preferences.
Fixation and embedding of cells for TEM can be very time-consuming and standard protocols may take several days from harvesting of the culture to the preparation of ultrathin sections and their analysis in the electron microscope. This process can be greatly accelerated by the use of microwave irradiation for fixation, dehydration and infiltration of cells 15, 16, 17, 18. Potassium permanganate (KMnO4) was one of the first fixatives used for TEM 19 and was soon later applied to the analysis of S. cerevisiae cells 20. It preserves lipid bilayers rather well and membrane-bounded organelles are well defined. However, at least with some protocols the resolution of fine ultrastructure may be less than optimal 11, mitochondrial morphology may not be well preserved and some small organelles and vesicles cannot be easily seen 21.
Here, we describe an optimized protocol for conventional permanganate fixation that we routinely use for the analysis of mitochondrial ultrastructure in yeast 22, 23, 24. Then we describe a novel protocol for microwave-assisted permanganate fixation of yeast, which allows to carry out fixation, dehydration, Epon infiltration and polymerization in less than 24 h and still preserves cellular ultrastructure well. We expect that both protocols will be useful for the analysis of mitochondria and other membrane-bounded organelles in yeast.
RESULTS
Conventional permanganate fixation of yeast
We describe a protocol for conventional permanganate fixation, based on the method published by Perkins and McCaffery 25 with some modifications 22. Cells were fixed with glutaraldehyde, which reacts with primary amino groups of lysines and irreversibly cross-links proteins 14. After washing, cells were post-fixed with KMnO4, which preserves membrane structure and provides some membrane staining 11, 14. After careful washing, cells were embedded in agar to facilitate handling of the sample. Then, the cell wall was permeabilized with sodium metaperiodate (NaIO4) 26, which breaks glycosylic bonds and thereby releases proteins from the cell wall, which makes it more permeable to viscous embedding solutions 11. This was followed by en bloc staining with uranyl acetate to enhance contrast of nucleic acids and proteins 11. Dehydration, Epon infiltration and contrast enhancement were performed essentially as described by Unger et al. 27 with minor modifications. The entire procedure is described in protocols 1 and 3 below.
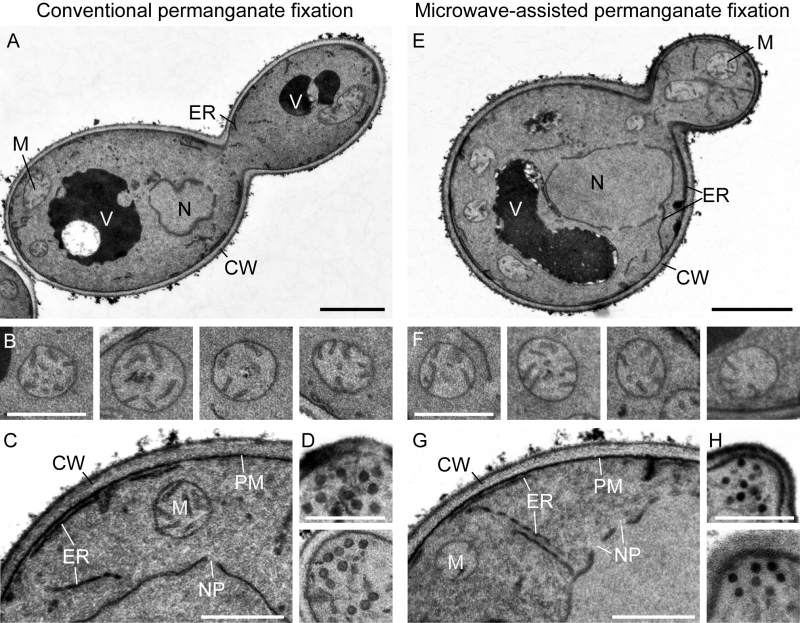
We found that this protocol is very useful to visualize organellar membranes in yeast cells (Figure 1A). Vacuoles appear electron-dense and/or electron-transparent, depending on their content. Mitochondrial cristae are intact and well resolved (Figure 1B). The ER is well resolved and the nuclear envelope is interrupted by nuclear pore complexes (Figure 1C). Secretory vesicles at the tips of small buds can be easily seen (Figure 1D).
–
Protocol 1: Conventional permanganate fixation of yeast
1. Grow yeast cells to logarithmic growth phase (OD600 ca. 0.3) and harvest 30 mL culture by centrifugation for 3 min at 1,600 xg.
2. Resuspend pellet in 10 mL freshly prepared 0.1 M sodium cacodylate, 1 mM CaCl2, 3% glutaraldehyde (EM grade), pH 7.2 and incubate for 1 h at room temperature.
3. Spin down cells, wash with 10 mL 0.1 M sodium cacodylate, 1 mM CaCl2, pH 7.2, centrifuge again, resuspend in 1 mL 0.1 M sodium cacodylate, 1 mM CaCl2, pH 7.2, transfer to a microfuge tube (2 mL), and centrifuge for 5 min at 800 xg in a microcentrifuge.
4. Resuspend cells in 1 mL freshly prepared 4% KMnO4 (in double distilled H2O, ddH2O) and incubate for 1 h at room temperature.
5. Wash cells four to six times by centrifugation for 5 min at 800 xg and careful resuspension in ddH2O until the supernatant is almost colorless. Remove supernatant.
6. Estimate the size of the pellet and add the same volume molten 2% agar noble (ca. 50°C) to the pellet, vortex thoroughly and briefly place the sample in ice water for rapid cooling and solidification of the agar.
7. Recover the sample from the tube, cut it into small pieces (about 1-2 mm edge length) and wash three times for 5 min in ddH2O.
8. Incubate up to ten agar blocs per sample in freshly prepared 0.5% NaIO4 (in ddH2O) for 15 min in a microfuge tube (2 mL) at room temperature and wash three times for 5 min in ddH2O.
9. Incubate agar blocs in 2% uranyl acetate overnight at room temperature in the dark and wash three times for 5 min in ddH2O.
10. Dehydrate the samples according to the following scheme. Use 2 mL of each solution per sample and incubate at 4°C: 30% ethanol, 15 min; 50% ethanol, 15 min; 70% ethanol, 15 min; 95% ethanol, 15 min; three times 100% ethanol, 20 min each; ethanol/propylene oxide mixture (1:1), 15 min; two times pure propylene oxide, 15 min each.
11. Perform Epon infiltration according to the following scheme. Use 2 mL of each solution per sample and incubate at room temperature: Epon/propylene oxide (1:3) for 3 h; Epon/propylene oxide (1:1) overnight; Epon/propylene oxide (3:1) for 3 h; pure Epon for 3 h; change Epon and incubate overnight.
12. To prepare for Epon polymerization, transfer the samples to an embedding mold, cover them with pure Epon and incubate for at least 12 h at 60°C.
Proceed with trimming, sectioning and contrast enhancement (protocol 3).
Microwave-assisted permanganate fixation of yeast
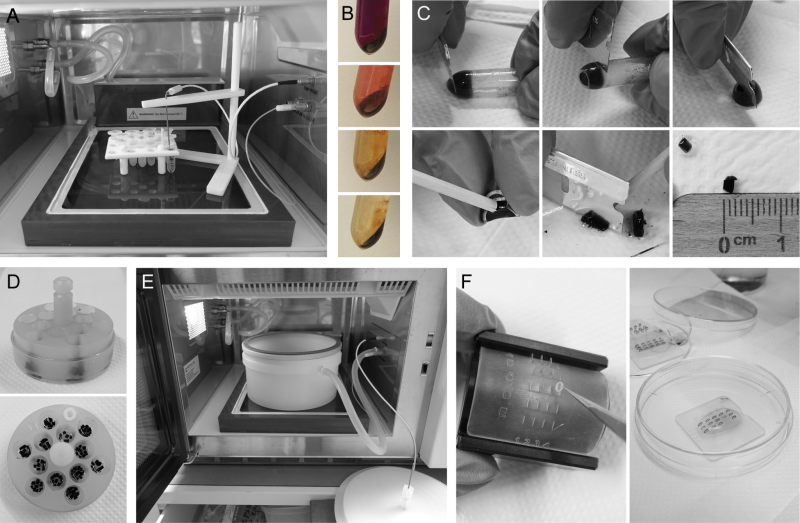
A similar regime was used for microwave-assisted permanganate fixation. We found that en bloc staining with uranyl acetate was not necessary and omitted this step. Embedding in agar was carried out after NaIO4 treatment, i.e. directly before dehydration and Epon infiltration, and acetone was used for dehydration. Time-consuming incubation steps during fixation, dehydration and Epon infiltration were replaced by short microwave treatments 28, 29. This reduces the time required for the entire procedure to the length of a working day. We find it convenient to carry out the final Epon polymerization step overnight and proceed with trimming and sectioning the next day. The procedure is described in protocols 2 and 3 below and illustrated in Figure 2.
–
We found that microwave-assisted permanganate fixation of yeast cells yields very similar results compared with the conventional fixation method. The membranes show good contrast and organellar structures, including mitochondria, ER, nuclear envelope and transport vesicles, are well preserved (Figure 1E-H).
Protocol 2: Microwave-assisted permanganate fixation of yeast
1. Grow yeast cells to logarithmic growth phase (OD600 ca. 0.3) and harvest 30 mL culture by centrifugation for 3 min at 1,600 xg.
2. Resuspend pellet in 600 µL freshly prepared 0.1 M sodium cacodylate, 1 mM CaCl2, 3% glutaraldehyde (EM grade), pH 7.2, transfer the sample to a microfuge tube (2 mL), subject cells to microwave treatment for 40 s, 750 W, max. 30°C, incubate for 5 min at room temperature and repeat microwave treatment for 40 s, 750 W, max. 30°C (Figure 2A).
3. Spin down cells for 3 min at 800 xg and wash two times with 0.1 M sodium cacodylate, 1 mM CaCl2, pH 7.2.
4. Resuspend cells in 600 µL freshly prepared 4% KMnO4 (in ddH2O), subject cells to microwave treatment for 40 s, 750 W, max. 30°C, spin down cells for 3 min at 800 xg, resuspend them in 600 µL 4% KMnO4, repeat microwave treatment for 40 s, 750 W, max. 30°C.
5. Wash cells four to six times by centrifugation for 5 min at 800 xg and careful resuspension in ddH2O until the supernatant is almost colorless (Figure 2B). Remove supernatant.
6. Resuspend cells in 600 µL freshly prepared 0.5% NaIO4 (in ddH2O), incubate for 15 min at room temperature and wash three times with 1 mL ddH2O.
7. Estimate the size of the pellet and add the same volume molten 2% agar noble (ca. 50°C) to the pellet, vortex thoroughly and briefly place the sample in ice water for rapid cooling and solidification of the agar.
8. Recover the sample from the tube, cut it into small pieces (about 1-2 mm edge length) and wash with ddH2O (Figure 2C).
9. For dehydration, transfer up to ten agar blocks per sample to the flow through basket (e.g. PELCO Prep-Eze 6 or 12 well specimen holder, Figure 2D) placed in a small Petri dish (5 cm diameter) filled with 50% acetone (the samples should be completely covered) and subject it to microwave treatment for 40 s, 750 W, max. 37°C. Repeat microwave treatment with increasing concentrations of acetone: 70%, 95%, 100%, 100%.
10. For Epon infiltration, transfer the flow through basket to a small Petri dish filled with acetone/Epon (1:1), place it into the vacuum chamber (Figure 2E) and subject it to microwave treatment for 5 min, 450 W, max. 42°C, 20”Hg vacuum (i.e. -67.7 kPa from atmospheric pressure). Repeat microwave treatment two times in pure Epon. Change the Petri dish with Epon between microwave treatments to avoid overheating of the samples and premature polymerization of pure Epon.
11. To prepare for Epon polymerization, transfer the samples to an embedding mold, cover them with pure Epon and incubate for at least 12 h at 60°C.
Proceed with trimming, sectioning and contrast enhancement (protocol 3).
Contrast enhancement
We used a standard protocol for contrast enhancement with uranyl acetate and lead citrate 30 as described by Unger et al. 27. It can be combined with both protocols for permanganate fixation.
Protocol 3: Contrast enhancement
1. After trimming, cut ultrathin sections of 60 to 70 nm and retrieve sections on Pioloform-coated copper grids.
2. Place the grid with the section down on a droplet of 2% uranyl acetate in a Petri dish and incubate for 15 to 20 min at room temperature in the dark. Wash the grid with the sections down in a series of three drops of ddH2O by incubation once for 1 min and two times for 2 min. Remove surplus water with filter paper. When processing many grids in parallel it is convenient to use a modified Hiraoka grid staining kit or similar for this and the following steps 31, 32 (Figure 2F).
3. Place the grid on a droplet of lead citrate 30 and incubate for 2 to 3 min. Wash in a series of three drops of ddH2O by incubation once for 1 min and two times for 2 min. Remove surplus water with filter paper.
4. Dry the grid for at least one hour before examination in the electron microscope. Grids can be stored indefinitely at room temperature.
Sample preparation without glutaraldehyde fixation or metaperiodate treatment
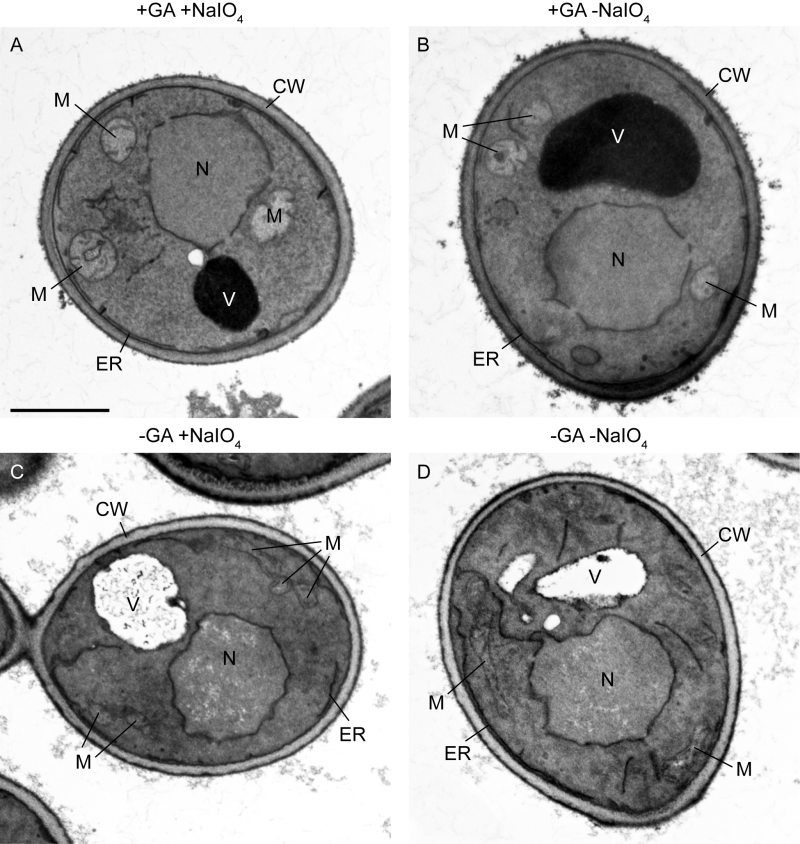
Some established permanganate fixation protocols lack glutaraldehyde fixation or metaperiodate treatment or both (see Table 1 and Discussion). Therefore, we asked whether these steps are necessary for optimal results of microwave-assisted permanganate fixation. To test this, we prepared the following four samples in parallel: The first sample was prepared according to the complete protocol 2 (Figure 3A); the second sample did not receive metaperiodate treatment, i.e. step 6 was omitted (Figure 3B); the third sample was not fixed with glutaraldehyde, i.e. steps 2 and 3 were omitted (Figure 3C); and the fourth sample was not fixed with glutaraldehyde and did not receive metaperiodate treatment, i.e. steps 2, 3 and 6 were omitted (Figure 3D). We observed that organellar ultrastructure was well preserved and membranes showed good contrast when cells were fixed with glutaraldehyde and only the metaperiodate treatment was omitted. When glutaraldehyde fixation was omitted the endomembrane system and nuclear envelope were still decently resolved, irrespective of whether or not samples were treated with metaperiodate. However, membrane contrast was worse compared with glutaraldehyde-fixed cells, mitochondrial ultrastructure was not well preserved and vacuoles appeared very transparent, presumably because most of their proteinaceous content was extracted. We conclude that glutaraldehyde fixation is critical for optimal preservation of organellar ultrastructure in microwave-assisted permanganate fixation. The metaperiodate treatment is optional, at least for the preparation of logarithmically grown wild type cells.
|
|
Glutaraldehyde |
KMnO4 |
NaIO4 |
Uranyl acetate* |
Resin |
|---|---|---|---|---|---|
|
Stevens and White, 1979 33 |
optional |
4% 2-4 h, 4°C |
– |
1% ON |
Spurr or Epon |
|
Erdmann et al., 1989 34 |
– |
1.5 % 20 min, RT |
– |
1% 2-12 h |
Epon |
|
Kaiser and Schekman, 1990 35 |
1% |
4% 2-4 h, 4°C |
– |
2% 12-18 h, 4°C |
Spurr |
|
Yaffe, 1995 36 |
3% 1 h, 4°C |
4% 1 h, 4°C |
– |
2% ON, 4°C |
Spurr |
|
Gammie and Rose, 2002 37 |
2% 30 min, RT |
4% 2-6 h, 4°C |
1% 15 min |
2% ON, 4°C |
LR White |
|
Perkins and McCaffery, 2007 25 |
3% 1 h |
4% 1 h, RT |
0.5% 15 min, RT |
2% ON, RT |
Spurr |
|
Griffith et al., 2008 21 |
– |
1.5% 2x 30 min, 4°C |
– |
– |
Spurr |
|
This study, protocol 1 |
3% 1 h, RT |
4% 1 h, RT |
0.5% 15 min, RT |
2% ON, RT |
Epon |
|
This study, protocol 2 |
3% MV |
4% MV |
optional |
– |
Epon |
* Some of these protocols use uranyl acetate for contrast enhancement after sectioning
MV, microwave treatment; ON, overnight; RT, room temperature
–
DISCUSSION
Potassium permanganate fixation is an established procedure for the analysis of yeast ultrastructure by TEM (reviewed in references 11, 13, 14). It preserves organelle structure rather well and yields a good membrane contrast. Numerous permanganate fixation protocols have been described in the literature and were successfully used to visualize organellar membranes (a few examples can be found e.g. in references 21, 25, 33, 34, 35, 36, 37; for an overview see Table 1). Yeast cells can be directly fixed with permanganate, dehydrated and embedded in Spurr’s resin 21. This fast and rather simple procedure is useful to reliably visualize the cell wall and most cellular membranes. However, it has been noted that mitochondrial morphology is not well preserved, small Golgi compartments, endosomes and transport vesicles are not easily seen 21 and fine ultrastructure may be lost to a certain extent 11. Consequently, cells are often first fixed with glutaraldehyde and then post-fixed with permanganate 14, 25, 33, 35, 36, 37. Although this appears not to be an essential step, some published protocols include a short sodium metaperiodate treatment of the cell wall to facilitate easier infiltration of the cell with embedding mixtures 25, 37. Also, fixed cells are frequently stained overnight with uranyl acetate before they are subjected to dehydration and embedding 25, 33, 34, 35, 36, 37. Different resins have been successfully used, including Epon 33, 34, Spurr’s 21, 25, 33, 35, 36 and LR White 37.
The protocol for conventional permanganate fixation described here allows rather easy handling of samples and does not require non-standard equipment. Microwave-assisted permanganate fixation reduces the time required for sample preparation starting from harvesting of the cells until Epon polymerization to less than 24 h. Samples produced with this method yield TEM images of the same quality compared with samples produced with conventional permanganate fixation. Therefore, it is a valuable method when a suitable microwave processing system is available and time constraints are critical or high throughput of samples is required. We suggest that cells should be first fixed with glutaraldehyde for optimal preservation of organellar ultrastructure and to obtain good membrane contrast during microwave-assisted permanganate fixation. Metaperiodate treatment supports efficient infiltration with Epon but may be omitted when this step is not critical.
There is a great variety of other methods for the preparation of yeast cells for electron microscopy, each of which has its specific advantages and limitations (reviewed in references 11, 13, 14, 21). In the following, we will briefly discuss two other methods that we have successfully used in the past. Osmium tetroxide (OsO4) is a post-fixative that stains and preserves membranes very well and is commonly used after initial fixation with glutaraldehyde and/or formaldehyde 11, 14. Osmium tetroxide is much less able to penetrate the cell wall than permanganate, and metaperiodate treatment is not sufficient to enable infiltration of cells 14. Instead, the cell wall is partially or completely removed by enzymatic digestion 12, 38. In our hands, protocols based on glutaraldehyde fixation, cell wall digestion with zymolyase, post-fixation with osmium tetroxide, en bloc staining with uranyl acetate and Epon embedding 27, 38 turned out to be very useful for the analysis of mitochondrial cristae architecture (see e.g. references 8, 39, 40, 41). We find that osmium tetroxide gives an even better resolution of fine ultrastructural details than permanganate and is well suited for electron tomography 8, 42. However, the procedure is rather time-consuming and zymolyase treatment has to be empirically optimized for each strain and growth condition as suboptimal cell wall digestion will result in insufficient ultrastructural preservation 14, 27.
The Tokuyasu method combines mild formaldehyde/glutaraldehyde fixation with infiltration of cells with cryoprotectants, freezing in liquid nitrogen and cryosectioning followed by imaging at room temperature 43. This method provides negative contrast and yields high resolution of membranes. A major advantage over chemical fixation with potassium permanganate or osmium tetroxide is that the antigenicity of proteins is preserved and therefore immunolabeling is possible 11, 21, 27, 44. According to our experience, protocols based on formaldehyde/glutaraldehyde fixation, metaperiodate treatment, sucrose cryoprotection (sometimes in combination with polyvinylpyrrolidone), liquid nitrogen freezing and cryosectioning 21, 27, 44 turned out to be very useful for the analysis of mitochondrial ultrastructure (see e.g. references 45, 46, 47). The Tokuyasu method gives a very good resolution of fine structural details and is well suited for electron tomography 47. It is similarly fast as microwave-assisted permanganate fixation. However, it requires a cryo-ultramicrotome and may be technically more challenging than chemical fixation methods.
MATERIALS AND METHODS
Growth and handling of yeast cells
S. cerevisiae strain BY4741 48 was grown to logarithmic growth phase at 30°C in liquid YPGal rich medium (1% yeast extract, 2% peptone, 2% galactose). Galactose was chosen as a fermentable carbon source that does not induce glucose repression 49 and therefore does not repress mitochondrial cristae biogenesis. Handling of yeast was according to standard procedures 50. Eppendorf (Hamburg, Germany) centrifuges 5804 R with rotor A-4-44 and 5415 D with rotor F45-24-11 were used for harvesting and washing of yeast cells and sample preparation. To facilitate handling of samples after fixation, cells were embedded in agar noble (BD Difco; reference number 214230; Becton, Dickinson and Company, Sparks, MD, USA).
Epon resin
The embedding resin was an Epon-812 51 replacement (termed Epon here). It was prepared according to standard procedures as described by Unger et al. 27, 13. It consisted of the following four components: 2-dodecenylsuccinic acid anhydride (DDSA) (Serva, Heidelberg, Germany, cat. no: 20755.01), glycid ether 100 (Serva, cat. no: 21045.02), methylnadic anhydride (MNA) (Serva, cat. no: 29452.02), 2,4,6-tris(dimethylaminomethyl)phenol (DMP-30) (Serva, cat. no: 36975.03). For EponA, 75.64 g glycid ether 100 was mixed with 95 g DDSA in a disposable plastic beaker. For EponB, 122 g glycid ether 100 was mixed with 115.7 g MNA in a separate beaker. Both beakers were covered with parafilm and stirred at room temperature until mixtures were homogeneous. Then, 150 g EponA was mixed with 220 g EponB. The beaker was covered with parafilm and stirred at room temperature to obtain a homogenous mixture. Finally, 6.5 mL of the catalyst EponC (DMP-30) was added while stirring. As soon as the solution adopted a homogenous yellowish to brownish color, 10 mL aliquots were prepared and stored at -20°C.
Microwave setup
A PELCO BioWave Pro+ (Ted Pella, Redding, CA, USA) microwave processing system was used for microwave treatment. It was equipped with a temperature probe with stand, PELCO ColdSpot Pro temperature control device and a PELCO EM Pro microwave vacuum chamber with a top plate with temperature probe port. For handling of samples, we used polypropylene Petri dishes (5 cm diameter), PELCO Prep-Eze 6 and 12 well specimen holder and microwave microcentrifuge tube holder (all Ted Pella). Final Epon polymerization was carried out overnight in an embedding mold without microwave treatment.
Transmission electron microscopy
Electron micrographs were taken with a JEOL JEM-1400 Plus transmission electron microscope operated at 80 kV, a JEOL Ruby CCD camera (3296×2472 pixels) and the TEM Center software Ver.1.7.19.2439 (JEOL, Tokyo, Japan). Adobe Photoshop CS6 (Adobe Inc., San Jose, CA, USA) was used for linear adjustments of brightness and contrast.
ACKNOWLEDGMENTS
We thank Rita Grotjahn for technical assis-tance. This work was supported by the Elitenetzwerk Bayern through the Biological Physics program and grants of the Deutsche Forschungsgemeinschaft (project numbers 433461293 to B.W. and 459304237 to T.K. and DFG Research Unit FOR2092 project number 239484859 to S.G.). Publication of this article is funded by the Open Access Publishing Fund of the University of Bayreuth.
COPYRIGHT
© 2024

Microwave-assisted preparation of yeast cells for ultrastructural analysis by electron microscopy by Mayer et al. is licensed under a Creative Commons Attribution 4.0 International License.