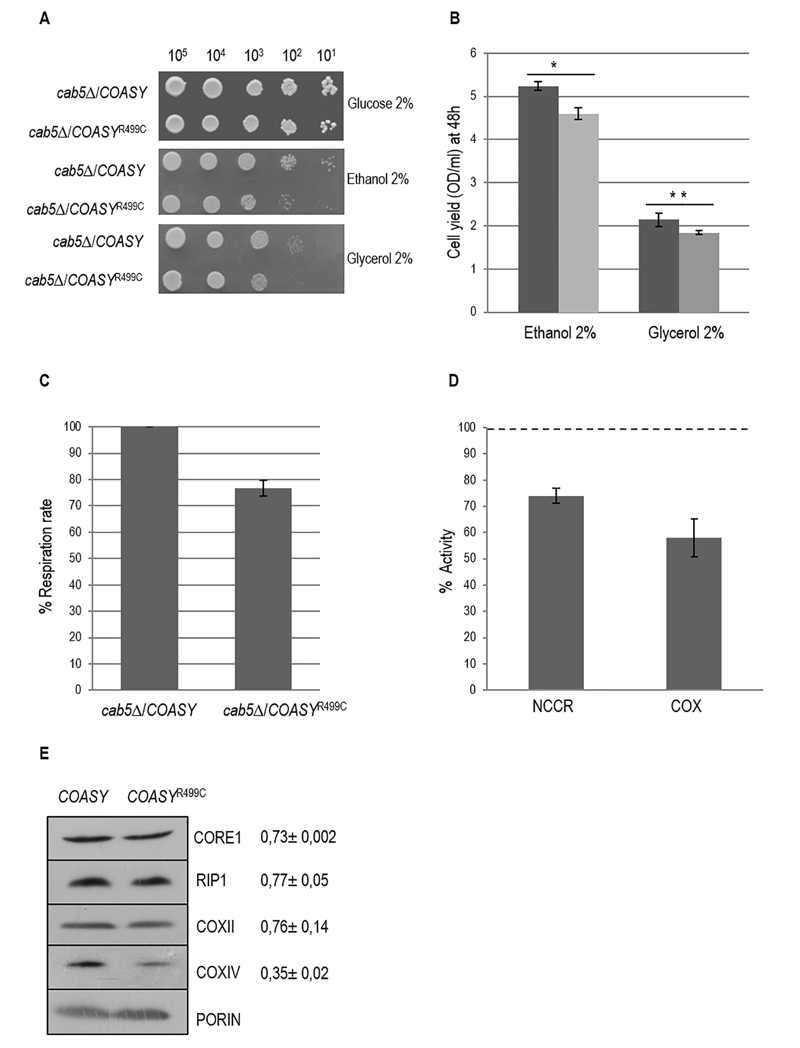
FIGURE 2: Characterization of mitochondrial functions.
(A) Oxidative growth phenotype. The strain W303-1B cab5Δ was transformed with a pYEX-BX plasmid carrying the wild-type COASY or the mutant allele COASYR499C. Equal amounts of serial dilutions of cells from exponentially grown cultures (105, 104, 103, 102, 101) were spotted onto mineral medium (40) plus 2% glucose, 2% ethanol or 2% glycerol without pantothenate. The growth was scored after 5 days of incubation at 28°C.
(B) Cell yield. Cell yield was calculated by growing cells on liquid medium containing ethanol or glycerol and measuring the optical density at 600 nm after 48h of growth (COASY black columns and COASYR499C grey columns). Values are mean of three independent experiments. * P < 0.05; **P < 0.01 (unpaired two-tailed t-test).
(C) Oxygen consumption rates. Respiration was measured in cells grown in mineral medium (40) plus 0.2% glucose and 2% galactose without pantothenate at 28°C. The values observed for the COASY mutant strain are reported as percentage of the respiration obtained in cells expressing the wild-type COASY gene.
(D) NADH-cytochrome c oxidoreductase (NCCR) and cytochrome c oxidase (COX) activities were measured in mitochondria extracted from cells grown exponentially at 28°C in mineral medium (40) plus 0.2% glucose and 2% galactose without pantothenate. The values of the COASY mutant are expressed as percentage of the activities obtained in the wild type strain.
(E) Steady state level of cIII and cIV subunits in cells carrying the wild-type COASY and the mutant allele. The filter was incubated with specific antibodies against Core1, Rip1, CoxII, CoxIV and Porin. The signals were normalized according to the control signal (porin) and taken as 1.00 the signal of the cab5∆/COASY (wild-type) strain.