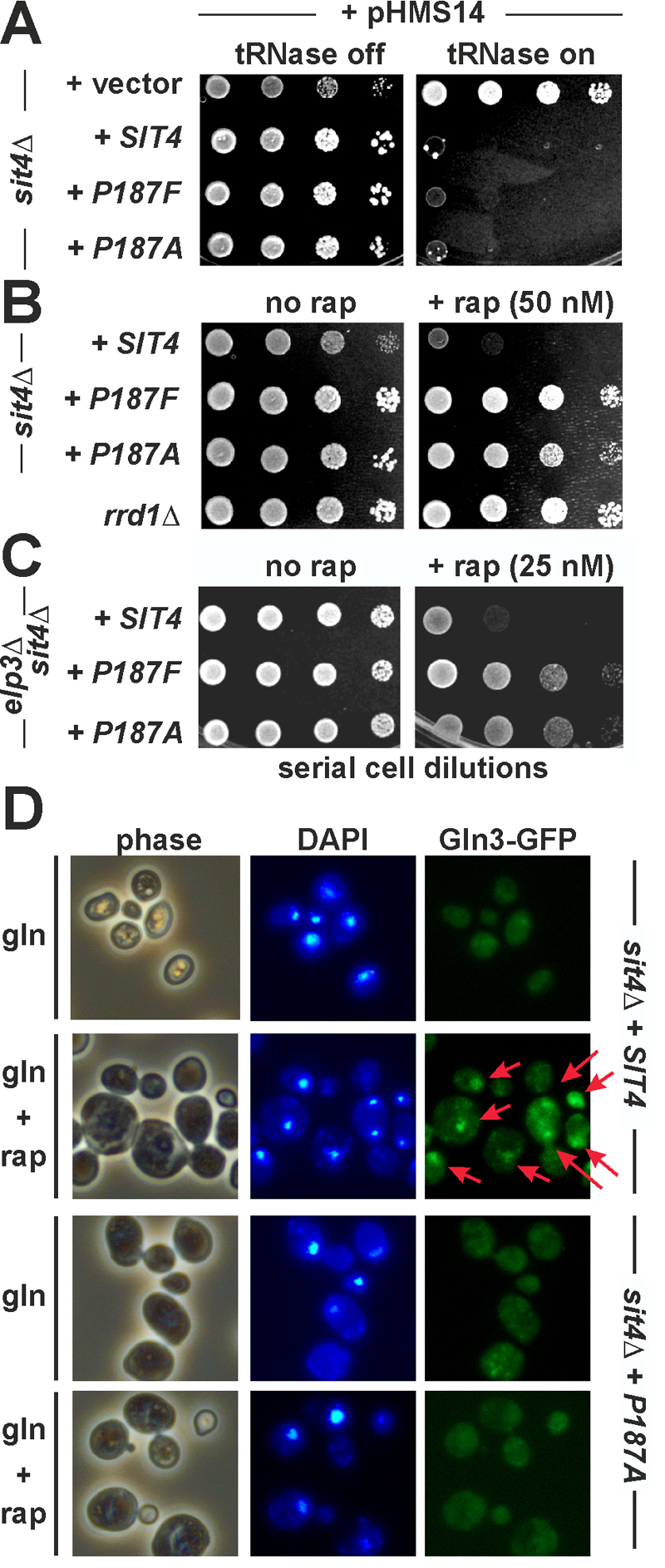
FIGURE 2: Substitutions of Sit4 proline residue 187 identify novel sit4 mutants that separate TOR-dependent phosphatase functions from TOR-insensitive ones.
(A) TOR-independent zymocin γ-toxin tRNase assay. The indicated sit4∆ backgrounds carrying empty vector, wild-type SIT4 and the P187F/A substitution alleles were transformed with the GAL1::γ-toxin expression vector (pHMS14) [12] and spotted onto glucose repressing (tRNase off) or galactose inducing (tRNase on) media. Growth was for 3 days at 30°C. Resistance or sensitivity towards tRNase toxicity is distinguished by growth or lack of growth, respectively.
(B) TOR-sensitive rapamycin phenotype. Ten-fold serial cell dilutions of a TOR signaling mutant (rrd1∆) and sit4∆ cells with genetic backgrounds as indicated in (A) were spotted onto medium containing rapamycin (+ rap, 50 nM) or no drug (no rap). Lack of growth indicates drug sensitivity, growth equals rapamycin resistance.
(C) The novel sit4 separation of function mutations suppress the sensitivity of Elongator mutants to rapamycin. A sit4∆elp3∆ double mutant with genetic backgrounds as indicated in (A) was grown on plates with no drug (no rap) or containing rapamycin (+ rap, 25 nM).
(D) Gln3 mislocalises as a result of the P187 substitution and fails to be imported into the nucleus under conditions of TOR inhibition. Cells carrying pRS416-GFP-Gln3 were grown in minimal medium containing glutamine (gln) as the sole N-source with or without 10 nM rapamycin (rap) and images taken in phase contrast, DAPI- and GFP-fluorescence modes (phase, DAPI, Gln3-GFP). Arrows indicate GFP signals and foci that co-localise with DAPI-stained nuclei.