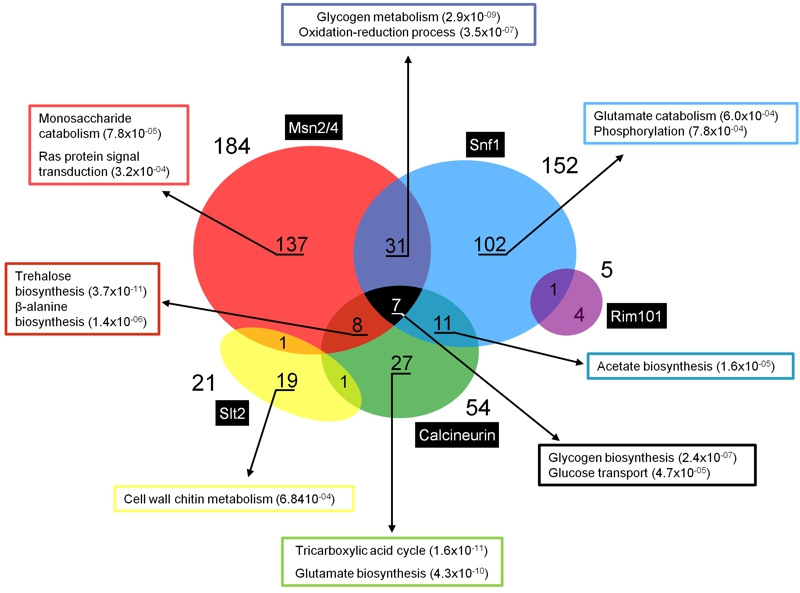
FIGURE 4: Genes regulated by multiple signaling pathways in response to alkaline pH stress.
Expression profiles have been collected from published transcriptomic data [18,22,36,56,68,97]. Gene Ontology analyses were based on FuncAssociate 2.0 [128] and YeastMine [129]. Numbers in parentheses denote the p-value for a given category.
18. Viladevall L, Serrano, R, Ruiz, A, Domenech, G, Giraldo, J, Barcelo, A, and Arino, J (2004). Characterization of the calcium-mediated response to alkaline stress in Saccharomyces cerevisiae. J Biol Chem 279(42): 43614-43624. http://www.ncbi.nlm.nih.gov/pubmed/?term=15299026
22. Serrano R, Martin, H, Casamayor, A, and Arino, J (2006). Signaling alkaline pH stress in the yeast Saccharomyces cerevisiae through the Wsc1 cell surface sensor and the Slt2 MAPK pathway. J Biol Chem 281(52): 39785-39795. http://www.ncbi.nlm.nih.gov/pubmed/?term=17088254
36. Ruiz A, Serrano R, and Arino J (2008). Direct regulation of genes involved in glucose utilization by the calcium/calcineurin pathway. J Biol Chem 283(20): 13923-13933. http://dx.doi.org/10.1074/jbc.M708683200
56. Casamayor A, Serrano R, Platara M, Casado C, Ruiz A, and Arino J (2012). The role of the Snf1 kinase in the adaptive response of Saccharomyces cerevisiae to alkaline pH stress. Biochem J 444(1): 39-49. http://dx.doi.org/10.1042/BJ20112099
68. Casado C, Gonzalez A, Platara M, Ruiz A, and Arino J (2011). The role of the protein kinase A pathway in the response to alkaline pH stress in yeast. Biochem J 438(3): 523-533. http://dx.doi.org/10.1042/BJ20110607
97. Lamb TM, Xu, W, Diamond, A, and Mitchell, AP (2001). Alkaline response genes of Saccharomyces cerevisiae and their relationship to the RIM101 pathway. J Biol Chem 276(3): 1850-1856. http://www.ncbi.nlm.nih.gov/pubmed/?term=11050096
128. Berriz GF, Beaver JE, Cenik C, Tasan M, and Roth FP (2009). Next generation software for functional trend analysis. Bioinformatics 25(22): 3043-3044. http://dx.doi.org/10.1093/bioinformatics/btp498
129. Balakrishnan R, Park J, Karra K, Hitz BC, Binkley G, Hong EL, Sullivan J, Micklem G, and Cherry JM (2012). YeastMine–an integrated data warehouse for Saccharomyces cerevisiae data as a multipurpose tool-kit. Database (Oxford) 2012 bar062-. http://dx.doi.org/10.1093/database/bar062