Research Reports:
Microbial Cell, Vol. 8, No. 2, pp. 57 - 64; doi: 10.15698/mic2021.02.742
A novel BR-SMAD is required for larval development in barber’s pole worm Haemonchus contortus
1 State Key Laboratory of Agricultural Microbiology, College of Veterinary Medicine, Huazhong Agricultural University, Wuhan, People’s Republic of China.
2 Department of Molecular Parasitology, Faculty of Life Sciences, Humboldt University, Berlin, Germany.
3 Department of Biological Sciences, Birla Institute of Technology and Science Pilani (BITS-P), Hyderabad, India.
Keywords: Haemonchus contortus, R-SMADs, BMP subfamily, RNAi, larval development.
Abbreviations:
AR-SMAD – activin/TGF-β/nodal activated R-SMAD,
BMP – bone morphogenic protein,
BR-SMAD – BMP and differentiation factor/Müllerian inhibiting substance activated R-SMAD,
Co-SMAD – common-mediator SMAD,
DBL1 – Dpp and BMP-like-1,
dsRNA – double strand RNA,
MH – mad homology domain,
NLS – nuclear localization sequence,
R-SMAD – receptor-regulated SMAD,
siRNA – small interfering RNA,
TGF-β – transforming growth factor β.
Received originally: 16/09/2020 Received in revised form: 07/12/2020
Accepted: 09/12/2020
Published: 23/12/2020
Correspondence:
Min Hu, State Key Laboratory of Agricultural Microbiology, College of Veterinary Medicine, Huazhong Agricultural University, Wuhan, People’s Republic of China; Tel: +086-13407126379; Fax: 86-27-87280408; mhu@mail.hzau.edu.cn
Conflict of interest statement: The authors declare no conflict of interest.
Please cite this article as: Fangfang Li, Peixi Qin, Lisha Ye, Nishith Gupta and Min Hu (2020). A novel BR-SMAD is required for larval develop-ment in barber’s pole worm, Haemonchus contortus. Microbial Cell 8(2): 57-64. doi: 10.15698/mic2021.02.742
Abstract
SMAD proteins mediate TGF-β signaling and thereby regulate the metazoan development; however, they are poorly defined in Haemonchus contortus–a common blood-sucking parasitic nematode of small ruminants. Here, we characterized an R-SMAD family protein in H. contortus termed HcSMA2, which is closely related to Caenorhabditis elegans SMA2 (CeSMA2) involved in the bone morphogenetic protein (BMP) signaling. Hcsma2 is transcribed in all developmental stages of H. contortus but highly induced in the adult male worms. The RNA interference with Hcsma2 retarded the transition of infective L3 into L4 larvae. Besides, the bimolecular fluorescence complementation revealed the interaction of HcSMA2 with a TGF-β-activated-R-SMAD (HcDAF8). Together these results show a BMP-like receptor-regulated SMAD in H. contortus that is required for larval differentiation and underscore an adaptive functional repurposing of BMP-signaling in parasitic worms.
INTRODUCTION
The known members of the TGF-β (transforming growth factor-β) superfamily can be classified into two main categories including the TGF-β/activin/nodal subfamily and the bone morphogenetic protein (BMP)/growth and differentiation factor (GDF)/Müllerian inhibiting substance (MIS) subfamily [1]. The classical SMAD-dependent TGF-β signaling involves the binding of an extracellular TGF-β homolog to a heterodimeric receptor complex, resulting in the activation and eventual nuclear translocation of specific cytoplasmic mediators (SMADs), which in turn regulate transcriptional activity by interacting with various transcription factors, co-activators and co-repressors [2]. In mammalian cells, such receptor-regulated SMADs (R-SMADs) are either activated by BMP type I receptors (aka BR-SMADs; SMAD1, SMAD5, SMAD8), or by activin and TGF-β type I receptors (aka AR-SMADs; SMAD2, SMAD3) [3]. TGF-β signaling also regulates the development of other metazoans including parasitic and free-living nematodes as described below.
–
Caenorhabditis elegans expresses five ligands of the TGF-β superfamily, of which DAF7 and DBL1 (Dpp and BMP-like-1), which belong to a canonical receptor-SMAD pathway, have been well studied. While DAF7 signaling is an important determinant of both dauer entry and exit decision [4], the DBL1 (Dpp and BMP-like-1) pathway regulates body size, male tail morphogenesis, cell lineage decision and innate immunity [5]. The DBL1 signal is relayed by a heterotetrameric receptor complex that is composed of two SMA6 type I and two DAF4 type II receptor subunits. The type II receptors first phosphorylate and activate the type I receptors, which in turn phosphorylate the downstream R-SMADs (SMA2 and SMA3). Phosphorylated R-SMADs interact with Co-SMAD (common-mediator SMAD; SMA4) and translocate to the nucleus for transcriptional regulation. Transcription factors that act with SMADs to exert DBL1-mediated responses include SMA9/Schnurri, LIN31/forkhead, and MAB31 [6]. Interestingly, the DBL1 and DAF7 pathways have some functional overlap in maintaining a non-dauer animal [7]. Both ligands (DBL1 and DAF7) interact with a sole type II receptor DAF4, and the specificity is determined by different type I receptors.
–
In Haemonchus contortus, previous work demonstrating the important roles of DAF7 signaling during larval development of this economically important nematode parasite has focused on TGF-β ligand [8], TGF-β type I receptor [9], TGF-β type II receptor [10]), TGF-β-regulated R-SMAD [11] and Co-SMAD [12]. Little is known about BMP (DBL1) signaling in this parasite, however. Here, we identified and characterized a new member of the H. contortus R-SMAD family (HcSMA2), which interacts with HcDAF8 (an R-SMAD) and contributes to the larval development.
RESULTS AND DISCUSSION
HcSMA2 is a member of the BMP-activated R-SMAD subfamily
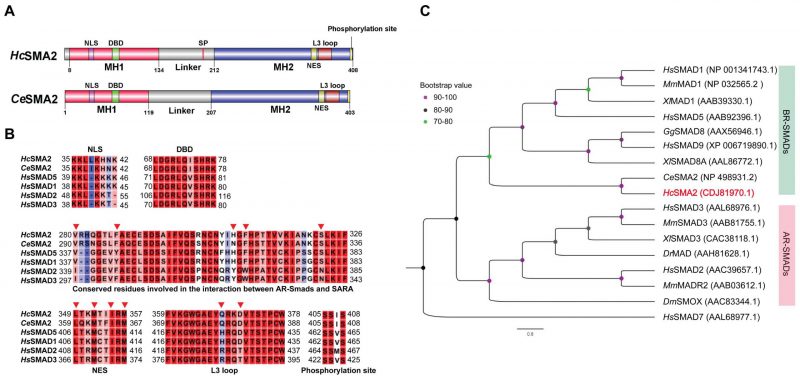
Our BLAST search revealed the presence of HcSMA2 protein in H. contortus, which shares 72% identity to CeSMA2. HcSMA2 (408 aa) contains conserved MH1 (mad homology domain; 8-134 aa) and MH2 (212-408 aa) domains along with signature motifs and residues of an R-SMAD (Fig. 1A). A nuclear localization signal and a DNA-binding β-hairpin domain were identified in the MH1 domain [13]. The proline-rich linker region contains an SP motif that may be phosphorylated by Serine/Threonine kinases [14]. Likewise, the MH2 domain harbors a conserved L3 loop, albeit Q368 and D371 that determine the interaction between R-SMADs and receptors diverge from that of BR-SMADs [15][16]. The VYHFS and IFYWN motifs regulate the interaction of R-SMADs and SARA (SMAD-anchoring for receptor activation) [17] in BR-SMADs and AR-SMADs, respectively. Similar to BR-SMADs, the VYHFS is broadly preserved in HcSMA2 (V280, H307, F309, S322; Fig. 1B). Specific residues (L349, M352, I355, M357) of the nuclear export signal (NES) could also be found in HcSMA2 [17]. Moreover, a typical SSxS motif for phosphorylation by type I receptor is present at the extreme carboxyl terminus (Fig. 1B), which is a hallmark of the R-SMAD subfamily [18]. Not least, the phylogenetic analysis revealed a clading of HcSMA2 and CeSMA2 along with other members of the BR-SMAD group (Fig. 1C).
–
–
HcSMA2 transcript is upregulated in the adult male worms
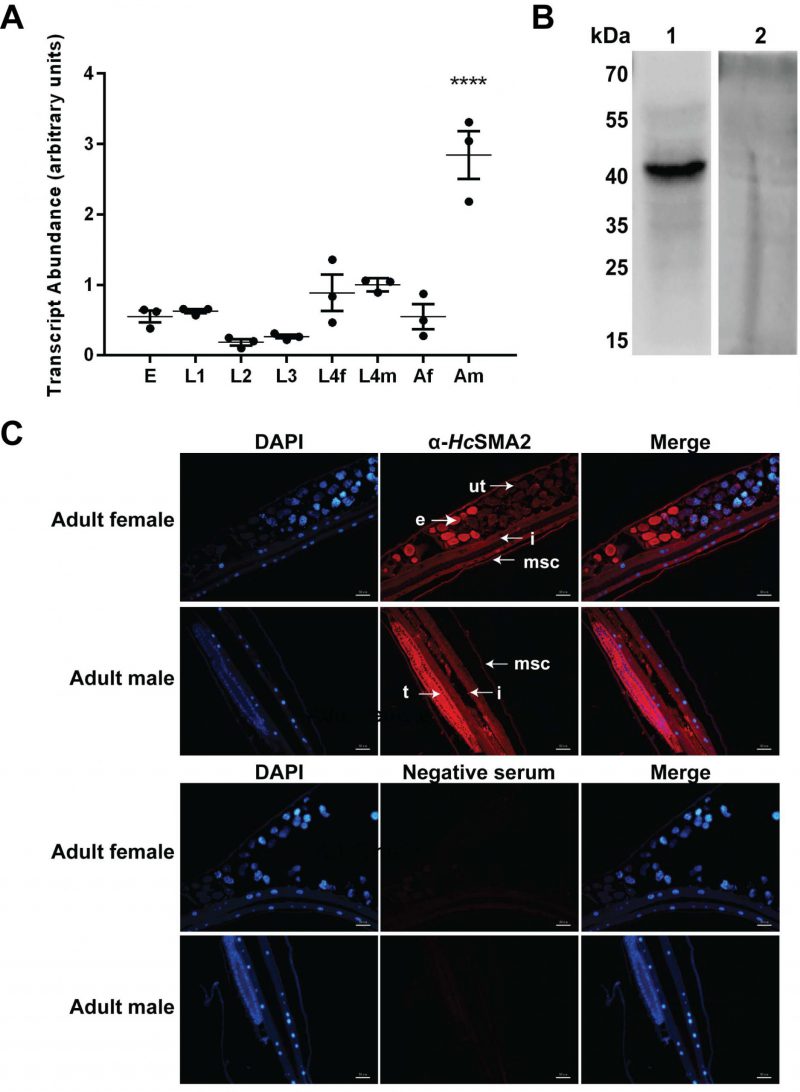
We examined the transcriptional profile of HcSMA2 during different developmental stages of H. contortus by RT-PCR and its tissue distribution in the adult worms by immunofluorescence assay (Fig. 2). Hcsma2 was transcribed in all stages with the highest transcript abundance in adult males (Fig. 2A). Next, we generated a polyclonal antibody against a synthetic partial polypeptide of HcSMA2, which recognized a band of the expected size (46-kDa) in the total parasite lysate (Fig. 2B). Immunofluorescence assay disclosed that HcSMA2 was expressed in the platymyrian muscle cells under the cuticle and in the intestinal cytoplasm of both female and male adults, as well as in eggs within the female worms, and in the testis of the male worms (Fig. 2C). The distribution of HcSMA2 contrasts with CeSMA2 in C. elegans, which is mainly expressed in the pharynx, reproductive system, and head cells of adult worms [19][20]. However, the overlapping expression of HcSMA2 and HcTGFBR2 [10] in the reproductive system and high transcript level of both in the adult male worms indicate that HcSMA2 may interact with HcTGFBR2 by conveying the BMP signals during spermatogenesis and embryonic development [21]. The expression of HcSMA2 in the platymyrian muscle cells and intestine also indicates its functions in muscle movement, invasion, and immunity [22].
–
–
HcSMA2, interacting with HcDAF8, is required for the worm development
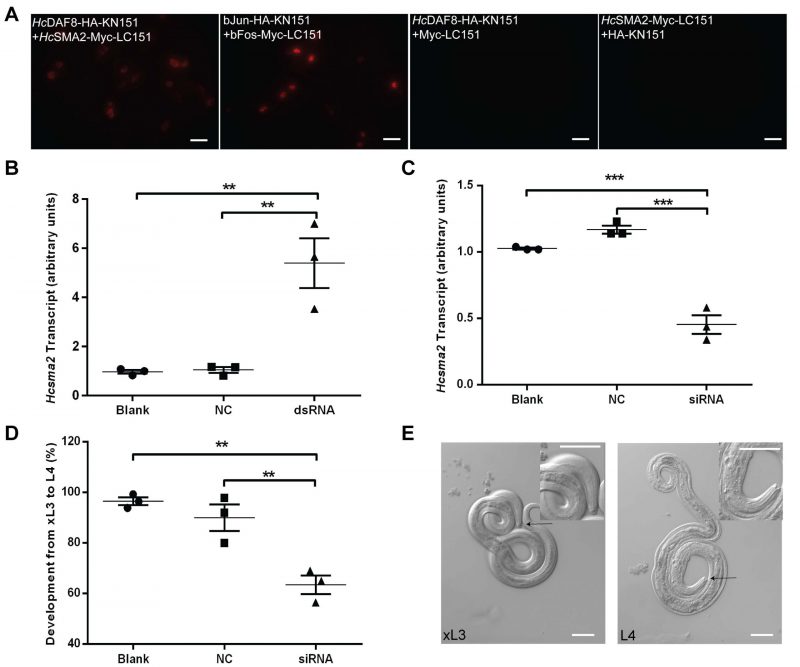
Our further work investigated whether HcSMA2 interacts with HcDAF8 [11] via a bimolecular fluorescence complementation assay (Fig. 3A). As a positive control, we transfected BHK21 cells with plasmid constructs encoding for bJun-HA-KN151 and bFos-Myc-LC151 fusion proteins, which exhibited a red signal in the nucleolus. Equally, cells co-transfected with HcSMA2-Myc-LC151 and HcDAF8-HA-KN151 constructs displayed red fluorescence in the cytoplasm. No fluorescence in cells harboring HcDAF8-HA-KN151 and Myc-LC151 proteins, or HcSMA2-Myc-LC151 and HA-KN151 protein pair was observed. These results indicated that HcSMA2 interacts with HcDAF8, and possibly regulate DAF7 signaling during the parasite development.
–
–
Last but not least, we examined the physiological importance of HcSMA2 in H. contortus by double strand RNA (dsRNA) or small interfering RNA (siRNA)-mediated RNA interference (Fig. 3B-E). Surprisingly, soaking of exsheathed L3 (xL3) larvae in dsRNA targeting the Hcsma2 caused a notable up-regulation of its transcript and did not exert a detectable phenotype (Fig. 3B). In contrary, Hcsma2 was silenced efficiently when the xL3 larvae were soaked in gene-specific siRNA (Fig. 3C), which resulted in a significant decline in morphogenesis to L4 larvae when compared to the control siRNA, and the blank control group (Fig. 3D). The morphological difference in the buccal capsule of xL3 and L4 larvae was also quite apparent, where the latter had its buccal capsule as an inverted cone and a pair of symmetric structures (mouthparts; Fig. 3E). These results suggest a role of HcSMA2 in differentiation of infective xL3 to L4 larvae, which is consistent with the function of the known members of the TGF-β and Insulin signaling [8][9][10][11][12][23]. In C. elegans, CeSMA2 regulates the body size [24], morphogenesis of the male worm's tail tip [25], as well as reproductive aging [21][26]. In contrast, HcSMA2 displays a notable functional divergence of the BMP-signaling. In conclusion, our work reveals a BMP-like receptor-regulated SMAD in H. contortus required for larval differentiation which has been repurposed to the parasitic lifestyle.
MATERIALS AND METHODS
Maintenance of H. contortus
The Haecon-5 strain of H. contortus was maintained by serial passages in experimental goats (12-14 weeks of age; raised helminth-free), as described previously [27]. Animals were infected orally with infective L3 stage larvae (7000 iL3s/kid). Eggs isolated from the goat feces by sucrose flotation [28] were cultured in a nutritive medium (0.1 mL of 1x Earle's balanced salt solution (Sigma-Adrich) and 0.5% (w/v) yeast extract) at 28°C [29]. The L1, L2, and iL3 stages were obtained from eggs cultured at 28°C for 1, 4 and 7 d, respectively [30]. The L4 stages and adults of both sexes were collected from the abomasum of the infected goats euthanized 8 and 30 d post-inoculation with iL3s [27]. Males and females of the L4 and adult stages were separated as reported earlier [31]. All experimental animals used in this project were treated in accordance with the protocol approved by the Ethics Committee of Huazhong Agriculture University, Wuhan, People's Republic of China (HZAUGO-2016-007).
–
Isolation of HsSMA2 in H. contortus
Homologs of CeSMA2 (WormBase, WS271, code ZK370.2) were identified in H. contortus (PRJEB506, PRJNA205202) (https://wormbase.org/tools/blast_blat) [32]. Using genomic and transcriptomic datasets for H. contortus[33], the coding sequence of HcSMA2 was isolated (GenBank: HF957107.1). In brief, the adult worms were homogenized, lysed with the TRIpure reagent (Aidlab, Beijing), and total RNA was extracted according to the manufacturer. The quality and yield of RNA were verified by electrophoresis and spectrophotometry (NanoDrop Technologies, USA). First-strand cDNA synthesis was performed by the Maxima H Minus First-Strand cDNA Synthesis Kit (Thermo Scientific, USA). The open reading frame of HcSMA2 was amplified from the first-strand cDNA using HcSMA2-F and HcSMA2-R primers (Table S1).
–
Bioinformatic analysis
The protein sequence of HcSMA2 (GenBank: CDJ81970.1) was analyzed for the conserved residues and motifs using Pfam (www.sanger.ac.uk/Software/Pfam), PROSITE (http://prosite.expasy.org) databases and CLC genomics workbench. The nuclear localization signal (NLS) was predicted by cNLS Mapper (http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi). Phylogenetic clading with representative members of the SMAD family was performed using the Maximum Likelihood (ML) method with 1,000 bootstrap replications.
–
Preparation of HcSMA2 antibody and immunoblot analysis
A synthetic peptide antigen (CDLQSHHELKAID) corresponding to 94-105 aa of HcSMA2-MH1 domain was used to immunize rabbits (Willget Biotech Co. Ltd., Shanghai, China). The HcSMA2 antiserum was tested by Western blot analysis of total protein extracted from the adult worms using a commercial kit (BestBio, China). Proteins were resolved by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto an Immobilon®-PSQ membrane (Merck Millipore Ltd). The immunoblot membranes were blocked with blocking buffer (1% (w/v) BSA (BioFROXX, Guangzhou, China) in PBS, 20% Tween-20; PBST) for 6 h at 4°C, washed 3 times with PBST and incubated with the HcSMA2 antiserum (1:1000 in PBST) overnight at 4°C. Samples were washed 6x in PBST and subsequently incubated with an HRP-conjugated goat anti-rabbit antibody (1:1000, Beyotime Biotechnology, China) for 2 h at 37°C, followed by 5x additional washes. Immunodetection was performed by chemiluminescence (WesternBright ECL kit; K-12045-D10, Aibio, China), and images were acquired by ChemiDoc XRS+ system (Bio-Rad, USA).
–
Indirect immunofluorescent assay
Fresh adult worms were washed with PBS (pH 7.4) and fixed overnight using 4% paraformaldehyde in PBS at 4°C [34]. Samples were washed, dehydrated, immersed in paraffin wax, and sliced. Sections (4 μm) were incubated in 3% hydrogen peroxide for 10 min at room temperature (24°C) to quench the endogenous peroxidase activity. They were pre-blocked with BSA for 20 min at 37°C, and then probed overnight with the HcSMA2 antiserum (1:100 in PBST) at 4 °C. The “negative control” sections were probed with the pre-immune rabbit serum (same dilution). Following three washes (5 min each), sections were incubated with Alexa Fluor® 594 goat anti-rabbit antibody (1:3000, ThermoFisher Scientific, R37117) for 50 min at 37°C. Unbound secondary antibody was washed off 3x by PBST, and samples were incubated with 4′,6-diamidino-2-phenylindole (DAPI) for 5 min at 24°C to visualize nuclei. Subsequently, samples were washed and mounted for fluorescent imaging (Olympus, BX51, Japan).
–
Double-strand RNA (dsRNA)-mediated RNA interference
The gene-specific dsRNA was synthesized as described elsewhere [9]. A fragment encoding partial MH2 domain (630-1220 bp) was amplified using Hcsma2i-F1/R1 (tagged with a T7 promoter site in the forward direction, and HindIII restriction enzyme site in the reverse direction) and Hcsma2i-F2/R2 (tagged with a HindIII restriction site (forward) and a T7 promoter site (reverse)) primer pairs (Table S1). Plasmids were linearized by HindIII and examined by agarose electrophoresis to test the enzymatic cleavage. Linearized DNA was gel-purified using Axygen® AxyPrep PCR Clean-Up kit (Axygen, Hangzhou, China), and precipitated in 2 volumes of 100% ethanol and 0.1 volume of 3 M sodium acetate (-20°C for 7 h). The DNA pellet was collected by centrifugation, washed with 75% ethanol, air-dried, resuspended in sterile water, and measured by spectrophotometer (NanoDrop Technologies, USA). The eventual DNA preparation was used as the template for in vitro transcription reactions using the MEGAscript® T7 transcription kit (Ambion, USA). The complementary RNA strands were annealed by mixing equimolar amounts and heated to 90°C for 5 min, followed by a further 2 h incubation at 37°C. The quality and concentration of the dsRNA were measured by electrophoretic analysis and spectrophotometry. Two sequences of the cry1Ac gene from Bacillus thuringiensis(Btcry1Ac; GenBank: GU322939.1) designed to produce the complementary single RNA strands (negative control) were amplified using Btcry1Ac-F1/R1 and Btcry1Ac-F2/R2 primers (Table S1). This fragment showed no significant homology with any nematode sequence in public databases [9]. The synthesis of dsRNA of Btcry1Ac was completed as described above. The dsRNA was stored at -80°C (80 µg/tube).
–
Specific dsRNAs encapsulated in a liposome preparation were added to worms. In brief, iL3s (10,000 larvae/group) were exsheathed with 0.15% sodium hypochlorite/PBS at 38°C until most of the shed sheaths were visible under the microscope (ca. 30 min), followed by 6x washes with PBS and 4x washes in Earle's balanced salt solution (EBSS, Sigma; pH adjusted to 5.2). The exsheathed L3 (xL3) larvae were soaked in 80 μL of EBSS. The liposome formulation of dsRNA was prepared by gentle mixing in a final volume of 20 μL (1 μL of lipofectamine 2000 (Invitrogen), 1 μL of RNasin (Promega), 80 µg of dsRNA and the residual volume of EBSS), allowed to stand for 10 min at room temperature and then added to the larvae (final concentration of dsRNA, 1 µg/µL). The larvae were incubated for 24 h at 37°C under 20% (v/v) CO2. Approximately, 200 larvae of each group were transferred to fresh EBSS culture medium for further 6 d to assess their developmental level and the remaining larvae were washed separately in a 10 mL glass tube with PBS for 4 times. The sedimented worms (8,000g for 4 min) were frozen in Trizol (Aidlab, Beijing, China) at -80°C for nucleic acid preparation and transcript analysis. On the 7th day, the worms that developed to the L4 stage were counted according to the phenotype of the buccal capsule [35][36].
–
Small interfering RNA (siRNA)-mediated RNA interference
Three custom siRNA in different segments of Hcsma2 were synthesized based on the gene coding sequence (GenePharma, Shanghai, China) (Table S2). The “negative control” siRNA lacking any homology with H. contortus sequence in public databases was also synthesized (Table S2). All siRNA preparations were stored in DEPC (50 μM)-treated water at −20°C. The clean exsheathed L3 worms were soaked in 80 μL of EBSS. Lipofectamine 2000 (5 μL) was mixed gently with 8.8 of μL EBSS and allowed to incubate for 5 min at room temperature followed by additional incubation with 6 μL of siRNA (siRNA-136 + siRNA-625 + siRNA-1130, 1 μM each) and 0.2 μL RNasin (8 U) for 15 min. The liposome formulation of siRNA (20 μL) was added into the exsheathed L3 cultures with a final volume of 100 μL in each well. The negative and blank controls were set up with 6 μL of “negative control” siRNA (3 μM) or 6 μL EBSS, respectively. Larvae were incubated at 37°C, 20% CO2 for 3 d. Approximately, 200 larvae of each group were transferred to fresh EBSS culture medium with 5% fetal bovine serum (Gibco, China) for another 4 d to assess their development. The remaining larvae were subjected to RNA extraction. The L4 larvae were counted on the 7th day.
–
Real-time PCR
RT-PCR was employed to examine the relative transcriptional abundance of Hcsma2 in different developmental stages (eggs, L1s, L2s, iL3s, L4f, L4m, Af and Am) of H. contortus (Haecon-5 strain) using the primers rtHcsma2-F and rtHcsma2-R (Table S1). Briefly, RNA was isolated from collected samples using TRIpure Trizol (Aidlab, Beijing, China). 1 μg RNA of the each stage was used to make the first-strand cDNA using HiScript III-RT SuperMix (Vazyme, Nanjing, China, R323-01). RT-PCR (10 μL) was set up using the ChamQ Universal SYBR qPCR Master Mix (Vazyme, Nanjing, China, Q711-02) in a thermal cycler (QuantStudio 3, Thermo Fisher Scientific, USA) under the following conditions: 50°C/2 min and 95°C/30 sec for the first cycle, then 95°C/15 sec, 60°C/15 sec and 72°C/20 sec for 40 cycles. The dissociation curve was generated under the following conditions: 95°C/15 sec, 60°C/1 min, and 95°C/1 sec. Each sample was tested in triplicate, employing β-tubulin (Accession ID, M76493) as a reference gene and corresponding primers (Tubulin-F/R, Table S1) [37]. The mean threshold cycle (Ct) values were subjected to estimate the relative quantities with respect to L4f (L4f = 1) using the 2-ΔΔCt method [38]. Besides, the transcripts of Hcsma2 in the RNAi-treated and control worms were quantified using RT-PCR. Here, the Hc18S rRNA gene (primers in Table S1) was used as a control as reported [39]. Fold change expression of Hcsma2 after RNAi calculated by 2-ΔΔCt method. ΔΔCt = [(CtRNAi,Hcsma2)-(CtRNAi,Hc18S)]-[(CtBlank,Hcsma2)-(CtBlank,Hc18S)].
–
Bimolecular fluorescence complementation assay (BiFC)
The bimolecular fluorescence complementation assay was performed using two plasmids as described in a previous study [40]. The full-length cDNA of HcDAF8 [11] and HcSMA2 were amplified with the primers Hcdaf8-HA-F/R and Hcsma2-Myc-F/R, respectively (Table S1). The plasmids pbJun-HA-KN151 and pbFos-Myc-LC151 provided by Yifan Wang [40] were digested by NheI/XhoI and NheI/PvuI, respectively. PCRs or digested products were purified (D2500-02, Omega, Shanghai, China) and sequenced. The amplicon of Hcsma2 was cloned into pbFos-Myc-LC151 between NheI and PvuI (replacing bFos) to generate the pHcSMA2-Myc-LC151 vector. Equally, the amplicon of HcDAF8 was cloned into pJun-HA-KN151 between NheI and XhoI sites (replacing bJun) to generate the pHcDAF8-HA-KN151 construct by homologous recombination kit (ClonExpress Entry One-Step Cloning Kit, Vazyme, Nanjing, China). Plasmids were extracted from E. coli (Plasmid Mini Kit II, D6950-02, Omega, Shanghai, China).
–
To transfect BiFC constructs, BHK21 cells were grown to 70~80% confluency in 6-well plates. pHcDAF8-HA-KN151 and pHcSMA2-Myc-LC151 were co-transfected in a ratio of 1:1 (2.5 μg each) using Lipofectamine 2000 (Invitrogen). Transfected cells were incubated for 20 h (37°C, 5% CO2) and subsequently imaged (Olympus BX51, Japan; Excitation, 543 nm; Emission, 580-680 nm). As a positive control, constructs expressing bJun and bFos (pbJun-HA-KN151 and pbFos-Myc-LC151) were utilized, whereas pHcDAF8-HA-KN151 and pMyc-LC151 or pHcSMA2-Myc-LC151 and pHA-KN151 plasmid pairs served as the negative controls.
–
Statistical analysis
Statistical analysis was performed using one-way ANOVA and p-values are calculated by Tukey's test (*, p≤0.05; **, p≤0.01; ***, p≤0.001; ****, p≤ 0.0001).
REFERENCES
- Miyazawa K, Shinozaki M, Hara T, Furuya T, Miyazono K (2002). Two major Smad pathways in TGF-beta superfamily signalling. Genes Cells 7(12): 1191-204. 10.1046/j.1365-2443.2002.00599.x
- Massagué J (2012). TGFβ signalling in context. Nat Rev Mol Cell Biol 13(10): 616-630. 10.1038/nrm3434
- Miyazono K, Maeda S, Imamura T (2005). BMP receptor signaling: transcriptional targets, regulation of signals, and signaling cross-talk. Cytokine Growth Factor Rev 16(3): 251-263. 10.1016/j.cytogfr.2005.01.009
- Thomas JH, Birnby DA, Vowels JJ (1993). Evidence for parallel processing of sensory information controlling dauer formation in Caenorhabditis elegans. Genetics 134(4): 1105-1117. 10.1101/gad.7.8.1632
- Patterson GI, Padgett RW (2000). TGF beta-related pathways. Roles in Caenorhabditis elegans development. Trends Genet 16(1): 27-33. 10.1016/s0168-9525(99)01916-2
- Gumienny TL, Savage-Dunn C (2013). TGF-β signaling in C. elegans. WormBook. 10.1895/wormbook.1.22.2
- Liu T, Zimmerman KK, Patterson GI (2004). Regulation of signaling genes by TGFβ during entry into dauer diapause in C. elegans. BMC Dev Biol 4: 11. 10.1186/1471-213X-4-11
- He L, Liu H, Zhang BY, Li FF, Di WD, Wang CQ, Zhou CX, Liu L, Li TT, Zhang T, Fang R, Hu M (2020). A daf-7-related TGF-β ligand (Hc-tgh-2) shows important regulations on the development of Haemonchus contortus. Parasit Vectors 13(1): 326. 10.1186/s13071-020-04196-x
- He L, Gasser RB, Korhonen PK, Di WD, Li FF, Zhang HR, Li FC, Zhou YQ, Fang R, Zhao JL, Hu M (2018). A TGF-β type I receptor-like molecule with a key functional role in Haemonchus contortus development. Int J Parasitol 48(13):1023-1033. 10.1016/j.ijpara.2018.06.005
- He L, Gasser RB, Li TT, Di WD, Li FF, Zhang HR, Zhou CX, Fang R, Hu M (2019). A TGF-β type II receptor that associates with developmental transition in Haemonchus contortus in vitro. PLoS Negl Trop Dis 13(12): e0007913. 10.1371/journal.pntd.0007913
- Li FF, Gasser RB, Liu F, Shan JN, Di WD, He L, Zhou CX, Wang CQ, Fang R, Hu M (2020). Identification and characterization of an R-Smad homologue (Hco-DAF-8) from Haemonchus contortus. Parasit Vectors 13(1): 164. 10.1186/s13071-020-04034-0
- Di WD, Liu L, Zhang T, Li FF, He L, Wang CQ, Ahmad AA, Hassan M, Fang R, Hu M (2019). A DAF-3 co-Smad molecule functions in Haemonchus contortus development. Parasit Vectors 12(1): 609. 10.1186/s13071-019-3855-3
- Dennler S, Huet S, Gauthier JM (1999). A short amino-acid sequence in MH1 domain is responsible for functional differences between Smad2 and Smad3. Oncogene 18(8): 1643-1648. 10.1038/sj.onc.1202729
- Kretzschmar M, Doody J, Massague J (1997). Opposing BMP and EGF signalling pathways converge on the TGF-beta family mediator Smad1. Nature 389(6651): 618-622. 10.1038/39348
- Chen YG, Hata A, Lo RS, Wotton D, Shi Y, Pavletich N, Massague J (1998). Determinants of specificity in TGF-beta signal transduction. Genes Dev 12(14): 2144-2152. 10.1101/gad.12.14.2144
- Lo RS, Chen YG, Shi Y, Pavletich NP, Massague J (1998). The L3 loop: a structural motif determining specific interactions between SMAD proteins and TGF-beta receptors. EMBO J 17(4): 996-1005. 10.1093/emboj/17.4.996
- Wu G, Chen YG, Ozdamar B, Gyuricza CA, Chong PA, Wrana JL, Massague J, Shi Y (2000). Structural basis of Smad2 recognition by the Smad anchor for receptor activation. Science 287(5450): 92-97. 10.1126/science.287.5450.92
- Attisano L, Lee-Hoeflich ST (2001). The Smads. Genome Biol 2(8): Reviews3010. 10.1186/gb-2001-2-8-reviews3010
- McKay SJ, Johnsen R, Khattra J, Asano J, Baillie DL, Chan S, Dube N, Fang L, Goszczynski B, Ha E, Halfnight E, Hollebakken R, Huang P, Hung K, Jensen V, Jones SJM, Kai H, Li D, Mah A, Marra M, McGhee J, Newbury R, Pouzyrev A, Riddle DL, Sonnhammer E, Tian H, Tu D, Tyson JR, Vatcher G, Warner A, et al. (2003). Gene expression profiling of cells, tissues, and developmental stages of the nematode C. elegans. Cold Spring Harb Symp Quant Biol 68: 159-169. 10.1101/sqb.2003.68.159
- Hunt-Newbury R, Viveiros R, Johnsen R, Mah A, Anastas D, Fang L, Halfnight E, Lee D, Lin J, Lorch A, McKay S, Okada HM, Pan J, Schulz AK, Tu D, Wong K, Zhao Z, Alexeyenko A, Burglin T, Sonnhammer E, Schnabel R, Jones SJ, Marra MA, Baillie DL, Moerman DG (2007). High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLoS Biol 5(9): e237. 10.1371/journal.pbio.0050237
- Luo S, Kleemann GA, Ashraf JM, Shaw WM, Murphy CT (2010). TGF-β and insulin signaling regulate reproductive aging via oocyte and germline quality maintenance. Cell 143(2): 299-312. 10.1016/j.cell.2010.09.013
- Chisholm AD, Xu S (2012). The Caenorhabditis elegans epidermis as a model skin. II: differentiation and physiological roles. Wiley Interdiscip Rev Dev Biol 1(6): 879-902. 10.1002/wdev.77
- Di WD, Gasser RB, He L, Li FF, Liu XF, Zhou CX, Zhou YQ, Fang R, Zhao JL, Hu M (2020). A serine/threonine-specific protein kinase of Haemonchus contortus with a role in the development. FASEB J 34(2): 2075-2086. 10.1096/fj.201900888RR
- Watanabe N, Ishihara T, Ohshima Y (2007). Mutants carrying two sma mutations are super small in the nematode C. elegans. Genes Cells 12(5): 603-609. 10.1111/j.1365-2443.2007.01077.x
- Nelson MD, Zhou E, Kiontke K, Fradin H, Maldonado G, Martin D, Shah K, Fitch DHA (2011). A bow-tie genetic architecture for morphogenesis suggested by a genome-wide RNAi screen in Caenorhabditis elegans. PLoS Genet 7(3): e1002010. 10.1371/journal.pgen.1002010
- Luo S, Shaw WM, Ashraf J, Murphy CT (2009). TGF-beta Sma/Mab signaling mutations uncouple reproductive aging from somatic aging. PLoS Genet 5(12): e1000789. 10.1371/journal.pgen.1000789
- Li FC, Lok JB, Gasser RB, Korhonen PK, Sandeman MR, Shi D, Zhou R, Li X, Zhou YQ, Zhao JL, Hu M (2014). Hc-daf-2 encodes an insulin-like receptor kinase in the barber’s pole worm, Haemonchus contortus, and restores partial dauer regulation. Int J Parasitol 44(7): 485-496. 10.1016/j.ijpara.2014.03.005
- Zawadzki JL, Kotze AC, Fritz JA, Johnson NM, Hemsworth JE, Hines BM, Behm CA (2012). Silencing of essential genes by RNA interference in Haemonchus contortus. Parasitology 139(5): 613-629. 10.1017/S0031182012000121
- Rossanigo CE, Gruner L (1991). Accuracy of two methods for counting eggs of sheep nematode parasites. Vet Parasitol 39(1-2): 115-121. 10.1016/0304-4017(91)90067-6
- Nikolaou S, Hartman D, Presidente PJ, Newton SE, Gasser RB (2002). HcSTK, a Caenorhabditis elegans PAR-1 homologue from the parasitic nematode, Haemonchus contortus. Int J Parasitol 32(6): 749-758. 10.1016/s0020-7519(02)00008-5
- Veglia F (1916). The anatomy and life-history of Haemonchus contortus (Rud.). Vet Res 4: 347-500. Available at: http://hdl.handle.net/2263/10471
- Ma G, Wang T, Korhonen PK, Stroehlein AJ, Young ND, Gasser RB (2019). Dauer signalling pathway model for Haemonchus contortus. Parasit Vectors 12(1): 187. 10.1186/s13071-019-3419-6
- Schwarz EM, Korhonen PK, Campbell BE, Young ND, Jex AR, Jabbar A, Hall RS, Mondal A, Howe AC, Pell J, Hofmann A, Boag PR, Zhu XQ, Gregory T, Loukas A, Williams BA, Antoshechkin I, Brown C, Sternberg PW, Gasser RB (2013). The genome and developmental transcriptome of the strongylid nematode Haemonchus contortus. Genome Biol 14(8): R89. 10.1186/gb-2013-14-8-r89
- Skinner TM, Bascal ZA, Holden-Dye L, Lunt GG, Wolstenholme AJ (1998). Immunocytochemical localization of a putative inhibitory amino acid receptor subunit in the parasitic nematodes Haemonchus contortus and Ascaris suum. Parasitology 117 (Pt1): 89-96. 10.1017/s0031182098002777
- Mapes CJ (1969). The development of Haemonchus contortus in vitro. I. The effect of pH and pCO2 on the rate of development to the fourth-stage larva. Parasitology 59(1): 215-231. 10.1017/s0031182000069961
- Sommerville RI (1966). The development of Haemonchus contortus to the fourth stage in vitro. J Parasitol 52(1): 127-136. 10.2307/3276403
- Guo XL, Zhang HL, Zheng XP, Zhou QJ, Yang Y, Chen XQ, Du AF (2016). Structural and functional characterization of a novel gene, Hc-daf-22, from the strongylid nematode Haemonchus contortus. Parasit Vectors 9(1): 422. 10.1186/s13071-016-1704-1
- Pfaffl MW (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29(9): e45. 10.1093/nar/29.9.e45
- Kotze AC, Bagnall NH (2006). RNA interference in Haemonchus contortus: suppression of beta-tubulin gene expression in L3, L4 and adult worms in vitro. Mol Biochem Parasitol . 145(1): 101-110. 10.1016/j.molbiopara.2005.09.012
- Wang Y, Fang R, Yuan Y, Pan M, Hu M, Zhou Y, Shen B, Zhao J (2016). Identification of host proteins, Spata3 and Dkk2, interacting with Toxoplasma gondii micronemal protein MIC3. Parasitol Res 115(7): 2825-2835. 10.1007/s00436-016-5033-2
SUPPLEMENTAL INFORMATION
![]() Download Supplemental Information
Download Supplemental Information
ACKNOWLEDGMENTS
We thank Robin B. Gasser (The Uni-versity of Melbourne) for helpful discussion. This study was supported by the National Key Basic Research Pro-gram (973 program) of China (2015CB150300), the Na-tional Natural Science Foundation (31872462) and the Natural Science Foundation of Hubei Province (2017CFA020) to M.H.
COPYRIGHT
© 2020

A novel BR-SMAD is required for larval development in barber’s pole worm Haemonchus contortus by Li et al. is licensed under a Creative Commons Attribution 4.0 International License.