Back to article: A novel BR-SMAD is required for larval development in barber’s pole worm Haemonchus contortus
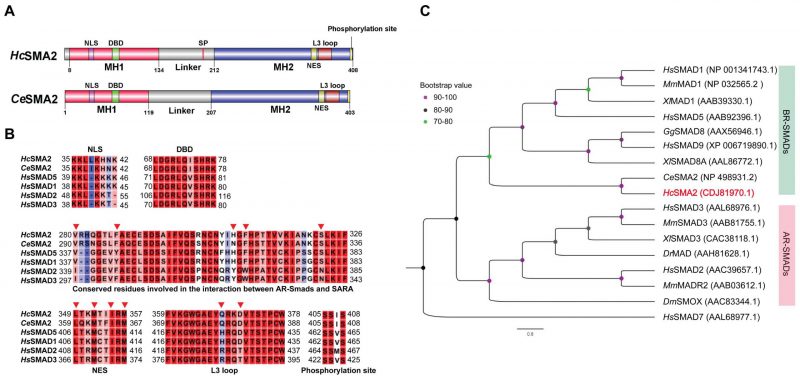
FIGURE 1: HcSMA2 displays conserved features of the BR-SMAD subfamily. (A) Primary structures of HcSMA2 and CeSMA2 with signature domains and motifs. (B) Sequence alignment of HcSMA2 with representative members of R-SMADs. The functional motifs include nuclear localization signal (NLS), DNA-binding β-hairpin domain (DBD), nuclear export signal (NES), L3 loop with R-SMAD subtype-specific amino acid residues, and phosphorylation motif. Residues involved in the interaction between R-SMAD and SARA as well as NES are highlighted (▾). The NCBI GenBank accession: HcSMA2, CDJ81970.1; CeSMA2, NP_498931.2; HsSMAD5, AAB92396.1; HsSMAD1, NP_001341743.1; HsSMAD2, AAC39657.1; HsSMAD3, AAL68976.1. (C) Phylogenetic analysis of HcSMA2 and selected homologs from Danio rerio, Gallus gallus, Homo sapiens, Mus musculus and Xenopus laevis, Drosophila melanogaster and C. elegans. The eventual cladogram was rooted using the inhibitory SMAD from H. sapiens SMAD7. Nodal support values for each clade are color-coded, and the accession numbers of individual proteins are listed in brackets next to each protein.