Research Reports:
Microbial Cell, Vol. 11, No. 1, pp. 235 - 241; doi: 10.15698/mic2024.07.828
Unresolved mystery of cyclic nucleotide second messengers, periplasmic acid phosphatases and bacterial natural competence
Department of Biology, University of Copenhagen, DK2200, Copenhagen, Denmark.
Keywords: cAMP, cGMP, cCMP, cUMP, natural competence, Haemophilus influenzae.
Received originally: 16/05/2024 Received in revised form: 21/06/2024
Accepted: 26/06/2024
Published: 18/07/2024
Correspondence:
Yong Everett Zhang, Department of Biology, University of Copenhagen, DK-2200 Copenhagen, Denmark; yong.zhang@bio.ku.dk
Conflict of interest statement: No conflict of interest is relevant with the contents of this article.
Please cite this article as: Kristina Kronborg, Yong Everett Zhang (2024). Unresolved mystery of cyclic nucleotide second messengers, periplasmic acid phosphatases and bacterial natural competence. Microbial Cell 11: 235-241. doi: 10.15698/mic2024.07.828
Abstract
We recently characterized the competitive inhibition of cyclic AMP (cAMP) on three periplasmic acid phosphatases, AphAHi, NadNHi, and eP4 (HelHi), in Haemophilus influenzae Rd KW20. This inhibitory effect is vital for orchestrating the nutritional growth and competence development in KW20. Initially discovered in Escherichia coli, the function of AphA remains however obscure. This study investigates the regulation of E. coli aphA expression under nutrient starvation conditions. Using transcriptional reporters with truncated aphA promoter sequences, we found that starvations of carbon and phosphate, but not amino acid, stimulated aphA expression through distinct promoter regions. Deletions of crp or cyaA abolished aphA expression, confirming their crucial roles. Conversely, CytR deletion increased aphA expression, suggesting CytR’s role as a repressor of aphA expression. Additionally, we extended the study of three other second messengers, i.e., cyclic GMP, cyclic UMP, and cyclic CMP, each sharing structural similarities with cAMP. Notably, cGMP competitively inhibits AphAHi’s acid phosphatase activity akin to cAMP. In contrast, both cUMP and cCMP stimulate AphAHi’s phosphatase activity in a concentration dependent manner. Collectively, these data imply a complicated connection between nucleotide metabolism, AphA, cyclic purine and pyrimidine nucleotides in bacterial nutrient uptake and natural competence.
INTRODUCTION
Cyclic AMP (cAMP) is a universal signaling molecule important to both eukaryotes and prokaryotes 1, 2 and proteins binding to cAMP are thus of high interest. Recently, we screened the Escherichia coli proteome and identified the non-specific acid phosphatase AphA as the other target protein of cAMP besides CRP 3. AphA is localized in the periplasmic space where it serves as a scavenging enzyme dephosphorylating a wide range of organic phosphomonoesters that otherwise cannot cross the inner membrane of E. coli
4, 5. Thereby, AphA allows E. coli to utilize these various phosphomonoesters for nutritional purposes and hence aid in bacterial survival. Similarly, AphA homolog in Salmonella typhimurium facilitates its uptake of external NAD 6.
We recently found that in Haemophilus influenzae, AphAHi, together with NadNHi and HelHi, serves to coordinate nutritional growth with the bacterium’s competence development 3. E. coli is, however, not naturally competent and AphA may play a different role in this bacterium. Previously, it was shown that the expression of aphA is induced by cAMP-CRP 7. In this study we aimed to identify the promotors responsible for aphA expression during different starvation conditions, to confirm the requirement of cAMP-CRP for aphA expression, and finally to identify regulators of its expression.
In the same study 3, we reported that cyclic GMP (cGMP) seems to affect AphA activity as well; however, the molecular effect of cGMP on AphA was not studied further. Furthermore, two additional cyclic nucleotides, namely the non-canonical second messengers cyclic CMP and cyclic UMP, have recently been acknowledged as authentic secondary messengers in bacteria 8. These molecules had long faced skepticism due to the absence of their dedicated synthetases 9. In bacteria, both cCMP and cUMP stimulate their specific effector proteins, mediating abortive phage infections 8. Given the similar chemical structures to cAMP, here we tested the effect of cGMP, cCMP, cUMP on the catalytic activity of H. influenzae AphAHi.
RESULTS
Carbon and phosphate starvations induce the transcription of E. coli aphAEc
The expression of E. coli aphAEc is stimulated by carbon starvation 7. To confirm this observation and to study how aphAEc transcription is regulated, we constructed four transcriptional reporter plasmids by cloning the serially truncated promoter regions (P1 to P4 from -433 to +30, Figure 1A-C) of aphAEc upstream of the lacZYA operon of the low-copy-number vector pGH254 previously used to analyze gene expression in E. coli 10. We first grew the E. coli strains with these reporters in MOPS synthetic rich medium and then shifted the cells to MOPS medium lacking either carbon, amino acid, or phosphate, to quantitate the transcription level of aphAEc. Consistent with previous report 7, carbon starvation led to an abrupt increase of aphAEc transcription from the promoter region P1 and P2, but moderately from P3, and not from P4 (Figure 1D), suggesting the -200 to -123 region is critical for aphAEc transcription during carbon starvation. A downshift of the reporter strains to low phosphate (from 1.32 mM to 0.067 mM) MOPS medium elicited a slow and moderate increase of aphAEc transcription from P2 and P3, but not from P4 or P1 (Figure 1E), suggesting that phosphate downshift stimulates the aphAEc transcription as well. At last, a downshift to MOPS medium without amino acid revealed no change of aphAEc transcription (Figure 1F), suggesting that amino acid starvation, and thus the stringent response, has no effect in aphAEc transcription. Of note, both P2 and P3 produced higher basal levels of aphA expression than P1 and P4 before the nutrient downshifts (Figure 1D-1F). Altogether, these data suggest that carbon and phosphate starvations induce the expression of aphAEc via overlapping but also distinct promoter regions.
–
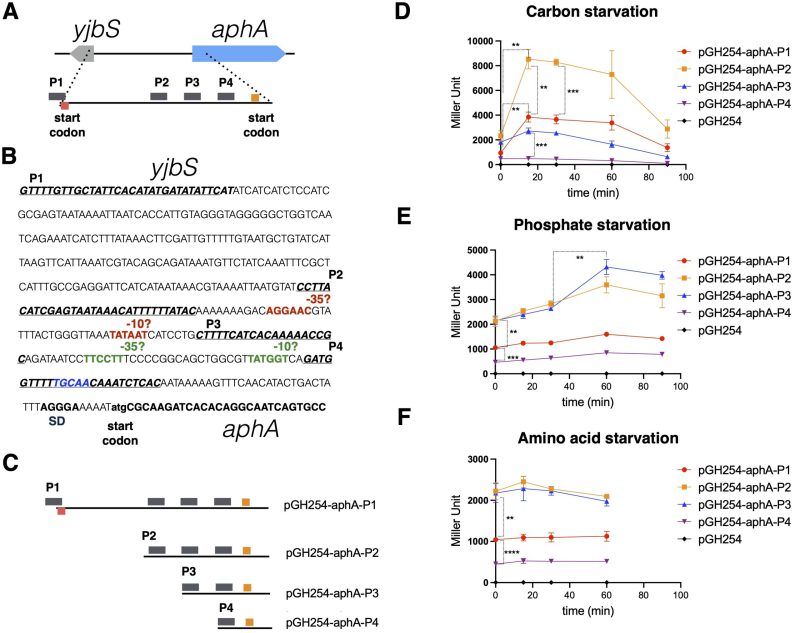 | FIGURE 1: Carbon and phosphate starvations stimulate the expression of E. coli aphA. (A) Schematic overview showing the promoter region of E. coli aphA gene, with the start and stop codons and potential four promoter sequences marked (in bars). (B) Sequence annotation of the potential promoter region of E. coli aphA. Text highlighted in blue is the CytR box; SD, Shine-Dalgarno sequence. (C) Schematic overview showing the promoter regions of aphA, which were transcriptionally fused to a lacZ reporter gene in the plasmid pGH254 [10]. (D, E, F) Beta-galactosidase assays of the various transcriptional reporters during starvations of carbon, phosphate, and amino acid. Time zero indicates the time point right before the downshift of cells. Three biological replicates were performed, and the average and standard deviations are shown. The significance of unpaired t-test is indicated by the symbols ** (p-value < 0.01), *** (p-value < 0.001), and **** (p-value < 0.0001). |
CyaA and CRP are essential for the aphAEc expression
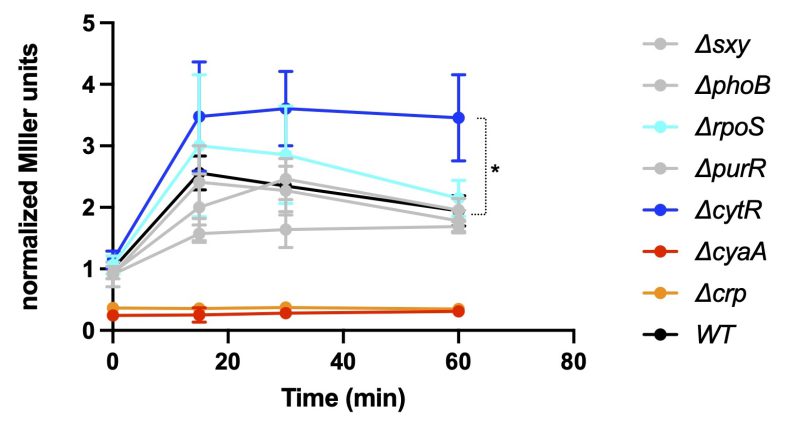
Carbon starvation in E. coli activates the cAMP-dependent CRP regulon. To confirm the link between carbon starvation and aphAEc transcription, we transformed the reporter plasmid pGH254-aphA-P2 into the KEIO deletion strains 11 of CRP or CyaA and performed a beta-galactosidase assay (see method). Besides, we included several other KEIO strains each deleted with a well-studied transcriptional regulator, i.e., Sxy, CytR, RpoS, PhoB, and PurR, given their (plausible) connections to bacterial nucleotide metabolism and AphA production in different organisms 12. Transcription of aphAEc dropped to basal levels in both ∆cyaA and ∆crp strains (Figure 2), demonstrating their essential roles in aphAEc expression upon carbon starvation. Additionally, in the ∆cytR mutant strain, transcription of aphA increased as compared to the wild type strain, suggesting that CytR suppresses the aphAEc transcription (see Discussion below). Indeed, inspection of the promoter region revealed a potential CytR box (Figure 1B, TGCAA, TTGC/tA) 13 within the P4 and N-terminal end of the aphAEc gene. The other mutant strains tested showed marginal, if at all, change of the aphAEc transcription. These data indicate that the production of AphAEc is controlled by both carbon and nucleotide metabolisms (see discussion below).
–
cUMP and cCMP stimulate, while cGMP inhibits, the acid phosphatase activity of AphA
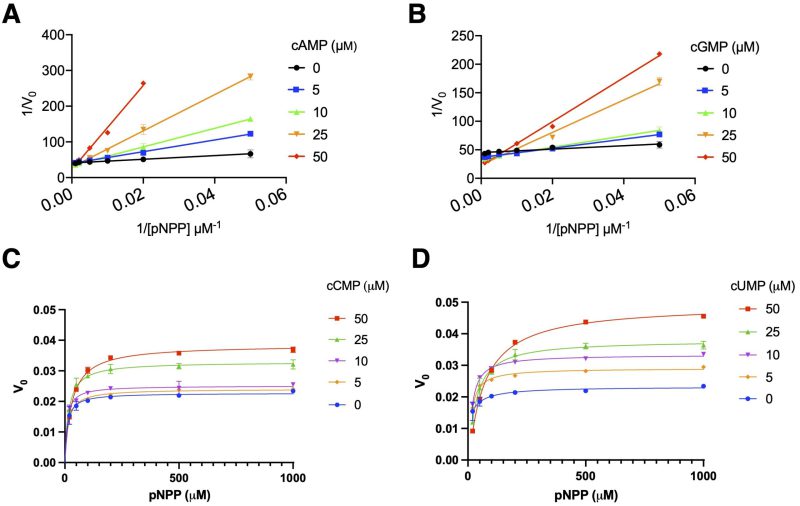
Our previous data 3 suggested that cGMP affects AphA acid phosphatase activity. Given the chemical similarities of cGMP, and cUMP and cCMP with cAMP, we tested their effects on AphAHi from Rd KW20. Figure 3B shows that cGMP appears to competitively inhibit AphAHi, like cAMP (Figure 3A), with the respective Ki values of 34.5 ± 5.2 and 6.4 ± 1.2 μM. Conversely, both cCMP and cUMP exhibit a concentration dependent stimulation of AphAHi phosphatase activity, with cUMP having an apparent higher efficacy than cCMP (Figure 3C, 3D). This intriguing discovery underscores the opposing effects of purine and pyrimidine cyclic nucleotides on AphAHi activity.
–
DISCUSSION
This short study first confirms the requirement for cAMP-CRP for aphA expression in E. coli 7. Further, it reveals CytR as a repressor of aphA expression (Figure 2). Instead, other tested regulators appear to exert only a minimal impact on aphA expression under carbon starvation. CytR typically represses gene expression by interacting with CRP and blocking the binding site of CRP 14. Upon binding to cytidine, the inducer, CytR unmasks the binding site of CRP, leading to derepressed gene expression. The discovery of a potential CytR box in the aphA promoter region (Figure 1B) is consistent with this notion and the observed increased expression of aphA (Figure 2). Cleavage of nucleotides by AphA releases nucleoside and phosphate, serving as carbon and phosphate sources respectively. The coupled stimulation of aphAEc transcription upon either carbon or phosphate downshift thus connects AphAEc to the nucleotide utilization in E. coli. Consistently, AphA contributes to the utilization of nucleotides (IMP, GMP) as the sole carbon source and IMP or inosine induced the expression of aphA in E. coli 4. These data suggest that AphAEc may be more involved in E. coli utilization of nucleotides as both carbon and phosphate sources (see below). Intriguingly, CytR is required for competence development in Vibrio cholerae 15. The introduction of external cytidine (100 mM) diminished V. cholerae competence to a level comparable to the deletion of cytRVc 16. On the other hand, purine, but not pyrimidine, nucleotides are involved in the competence development of H. influenzae 12, 17. Of note, H. influenzae has no CytR homolog and V. cholerae has no AphA homolog. Instead, E. coli encodes both CytR and AphA. Furthermore, E. coli encodes some homolog proteins involved in natural competence 18, 19, despite that the phenomenon or the right condition under which E. coli becomes competent has not been reported yet. Therefore, it is generally believed that the laboratory strain of E. coli has lost some key unidentified competence regulator. Consistently, an environmental E. coli strain ED1 20 was found to be naturally competent; however, the causative genomic differences remain undefined. Altogether, these observations concur with the notion that the natural competence system evolved from bacterial uptake of external DNA and nucleotides as nutrients 19, 21, 22.
We surprisingly found that both cCMP and cUMP stimulate the catalytic activity of AphAHi in contrast to the inhibitory effects of cGMP and cAMP (Figure 3). Interestingly, differential effects of cGMP and cUMP were observed before 23, and both cCMP and cUMP stimulate their specific effector proteins, mediating abortive phage infections 8. It thus appears that cUMP and cCMP indeed assume consistently a positive effect on their target proteins as compared to cAMP and cGMP. However, these cC/UMP specific cyclases (Pyrimidine nucleotide cyclase, Pyc) have a relatively narrow phylogenetic distribution, as noted by the authors 8. Additionally, many extensively studied model (pathogenic) bacteria, such as E. coli, H. influenzae, and V. cholerae, do not possess the Pyc enzymes, despite that some bacteria produce exotoxins e.g., ExoY from Pseudomonas aurogenosa 24 which produces cUMP (and cGMP) inside host cells. Furthermore, while host mammalian cells may produce cC/UMP 25, 26 (possibly through the promiscuous activities of cAMP and cGMP cyclases) and excrete them, their concentrations are anticipated to be lower than that of cAMP and cGMP. On the other hand, in certain microbial community, there is the intriguing possibility that extracellular cC/UMP may stimulate the activities of AphAHi (as well as NadNHi and HelHi), facilitating the nutritional uptake of NAD and nucleotides, ultimately impacting the growth of H. influenzae. This opens the possibility that the dynamic regulation of these three enzymes by various cyclic nucleotide molecules produced by KW20, other microbiota, and host cells collectively influences when and how KW20 becomes competent to uptake external DNA. This broader perspective on inter-species communication is potentially relevant and warrants further exploration. What are the mechanistic details of cU/CMP’s stimulatory effect on AphAHi? Why would cAMP inhibit the catalytic activity of AphA while simultaneously stimulating its transcription? Answering these outstanding questions may provide a better understanding of the conditions and mechanisms leading to the development of bacterial competence and drug resistance.
MATERIAL AND METHODS
Construction of transcriptional reporters of aphA-lacZ
Strains used in this study are listed in Table 1 and primers are listed in Table 2.
To make the APHA-1 transcriptional reporter, the primers pYZ262 and pYZ266 were used to amplify the P1 promoter region (Figure 1) of aphAEc and cloned into the plasmid vector pGH254 10 via the EcoRI and BamHI restriction sites. This places the P1 promoter region of aphAEc in front of the lacZYA genes on pGH254 resulting in the transcriptional reporter pGH254-APHA-1. Likewise, P2, P3 and P4 promoter regions of aphAEc were amplified using primers pYZ263 and pYZ266, primers pYZ264 and pYZ266, and primers pYZ265 and pYZ266, respectively, and similarly cloned into pGH254 to construct the pGH254-APHA-2, pGH254-APHA-3, pGH254-APHA-4, respectively.
Table 1. Bacterial strains used in this study.
|
Strain |
Relevant features |
Reference |
|---|---|---|
|
YZ254 |
Bl21 DE3, pET28a-his.aphAhi, Kan |
|
|
YZ351 |
TB28 pGH254-APHA-1, Kan |
This study |
|
YZ352 |
TB28 pGH254-APHA-2, Kan |
This study |
|
YZ353 |
TB28 pGH254-APHA-3, Kan |
This study |
|
YZ354 |
TB28 pGH254-APHA-4, Kan |
This study |
|
YZ356 |
TB28 pGH254, Kan |
This study |
|
YZ1047 |
Keio sxy::kan, pGH254-APHA-2, Kan, Cam |
This study |
|
YZ1048 |
Keio phoB::kan, pGH254-APHA-2, Kan, Cam |
This study |
|
YZ1049 |
Keio rpoS::kan, pGH254-APHA-2, Kan, Cam |
This study |
|
YZ1050 |
Keio purR::kan, pGH254-APHA-2, Kan, Cam |
This study |
|
YZ1051 |
Keio cytR::kan, pGH254-APHA-2, Kan, Cam |
This study |
|
YZ1052 |
Keio cyaA::kan, pGH254-APHA-2, Kan, Cam |
This study |
|
YZ1053 |
TB28 crp::tet, pGH254-APHA-2, Tet, Cam |
This study |
|
YZ1054 |
Keio WT, pGH254-APHA-2, Cam |
This study |
Table 2. Primers used in this study.
|
Name |
Seqquence |
|---|---|
|
pYZ262 |
GGAATTCGTTTTGTTGCTATTCACATATGATATATTC |
|
pYZ263 |
GGAATTCCCTTACATCGAGTAATAAACATTTTTTATAC |
|
pYZ264 |
GGAATTCCTTTTCATCACAAAAACCGC |
|
pYZ265 |
GGAATTCGATGGTTTTTGCAACAAATCTCAC |
|
pYZ266 |
CGGGATCCGGCACTGATTGCCTGTGTGA |
Beta-galactosidase assay
Overnight cultures of E. coli strains grown in MOPS medium (18 h, 37°C, 160 rpm) were back-diluted into fresh MOPS medium and grown to OD600=0.3-0.5 before they were shifted to a MOPS medium lacking either phosphate, glucose, or amino acids. During growth, pelleted cell samples were collected and immediately frozen until use. For beta-galactosidase assays, the collected sample cells were thawed and resuspended in a resuspension buffer composed of 100 mM Tris (pH 7.8), 32 mM Na3PO4 (pH 7.8), 1 mM PMSF and 19 mM β-mercaptoethanol and were sonicated (10% amplitude) until complete lysis was observed. Protein concentration was measured using the Bradford assay, and equal amounts of total protein from each lysed sample were resuspended in a reaction buffer composed of 76 mM Tris (pH 7.8), 24 mM Na3PO4 (pH 7.8), 0.03% SDS, 1.3 mM MgSO4, 0.4 mg/ml DNase I, 1 mM PMSF and 19 mM β-mercaptoethanol to a final volume of 75 μl. The β-galactosidase reaction was started by adding 60 μl 4 mg/ml ONPG to the reaction mixture and was terminated by the subsequent addition of 150 μl 1 M Na2CO3. Finally, the samples were spun down for 2 min at 14600 rpm and 200 μl supernatant was transferred to a clear flat-bottom 96-well plate (Greiner) and the absorbance was read at 420 nm in a plate reader. Three biological replicates each with two technical replicates were performed, and the average and standard deviations are shown.
AphA purification and enzymatic assays
Both the purification and enzymatic assay of AphA were performed in the same method as in 3. Briefly, E. coli BL21 DE3 strains containing the vector pET28a-his.aphAhi were grown up in LB medium to OD600=0.6-0.8, when 0.5-1 mM IPTG was added to induce the expression of His-AphAHi for 4 hours at 37°C. Harvested Cells were lysed via sonication (Branson) in cold lysis buffer (5% glycerol 50 mM Tris pH=7.6, 150 mM NaCl, 10 mM imidazole, 5 mM β-mercaptoethanol) containing cOmpleteTM Protease Inhibitor Cocktail. The cleared supernatant was then loaded on Ni-NTA resins on a Poly-Prep column. The resins were washed with cold wash buffer (5% glycerol 50 mM Tris pH=7.6, 150 mM NaCl, 20 mM imidazole), eluted with elution buffer (5% glycerol 50 mM Tris pH=7.6, 150 mM NaCl, 500 mM imidazole) and further purified via the ÄKTA system on a Superdex 200 Increase 10/300 GL column. The purity of purified protein was verified using SDS-PAGE gel and its concentration was quantified using Bradford assay. We then used the pNPP as the substrate to assess the enzymatic reaction of His-AphAHi. For this, 15 nM of His-AphAHi was used in reaction buffer containing 50 mM sodium acetate (pH 5.6), 0.1 M NaCl, 1 mM MgCl2, 0.01% Triton X-100 and varying concentrations of cAMP, cGMP, cCMP and cUMP. The enzymatic reaction was started by adding varied pNPP and stopped by transferring 60 μl of the reaction mixture into 100 μl 3 M NaOH in a 96 well plate (Greiner, flat bottom) at defined time points. The amount of reaction product was quantitated by the absorption at 405 nm. Two biological replicates each with two technical replicates were performed, and the average and standard deviations are shown.
Preparation of chemicals
cAMP (SIGMA, A9501) was dissolved in Milli Q (MQ) H2O. cUMP (BioLog, 56632-58-7) and cCMP (BioLog, 54925-33-6) were each dissolved in a buffer of 25 mM Tris-HCl (pH 7.4) and 100 mM NaCl to a concentration of 250 mM and were then diluted to the desired concentrations in MQ H2O. p-nitrophenyl phosphate (pNPP, SIGMA, S0942) was dissolved in 50 mM NaOAc to a concentration of 50 mM and was then serially diluted in the same buffer to the desired concentrations.
ACKNOWLEDGMENTS
We are grateful to Kenn Gerdes for useful discussions. This study was supported by a Danmarks Frie Forskningsfond grant (2032-00030B) and Novo Nordisk Foundation Project Grant (NNF19OC0058331) to Y.E.Z.
COPYRIGHT
© 2024

Unresolved mystery of cyclic nucleotide second messengers, periplasmic acid phosphatases and bacterial natural competence by Kronborg and Zhang is licensed under a Creative Commons Attribution 4.0 International License.