Reviews:
Microbial Cell, Vol. 1, No. 6, pp. 163 - 178; doi: 10.15698/mic2014.06.152
Cell-autonomous mechanisms of chronological aging in the yeast Saccharomyces cerevisiae
Department of Biology, Concordia University, Montreal, Quebec H4B 1R6, Canada.
# These authors contributed equally to this work.
Keywords: yeast chronological aging; systems biology of cellular aging; cell-autonomous mechanisms of longevity regulation; proteostasis; lipid metabolism; mitochondria; carbohydrate metabolism.
Received originally: 07/04/2014 Received in revised form: 11/05/2014
Accepted: 13/05/2014
Published: 27/05/2014
Correspondence:
Vladimir I. Titorenko, 7141 Sherbrooke Street, West, SP Building, Room 501-13; Montreal, Quebec H4B 1R6, Canada vladimir.titorenko@concordia.ca
Conflict of interest statement: The authors declare no conflict of interest.
Please cite this article as: Anthony Arlia-Ciommo, Anna Leonov, Amanda Piano, Veronika Svistkova and Vladimir I. Titorenko (2014). Cell-autonomous mechanisms of chronological aging in the yeast Saccharomyces cerevisiae. Microbial Cell 1(6): 163-178.
Abstract
A body of evidence supports the view that the signaling pathways governing cellular aging – as well as mechanisms of their modulation by longevity-extending genetic, dietary and pharmacological interventions – are conserved across species. The scope of this review is to critically analyze recent advances in our understanding of cell-autonomous mechanisms of chronological aging in the budding yeast Saccharomyces cerevisiae. Based on our analysis, we propose a concept of a biomolecular network underlying the chronology of cellular aging in yeast. The concept posits that such network progresses through a series of lifespan checkpoints. At each of these checkpoints, the intracellular concentrations of some key intermediates and products of certain metabolic pathways – as well as the rates of coordinated flow of such metabolites within an intricate network of intercompartmental communications – are monitored by some checkpoint-specific ʺmaster regulatorʺ proteins. The concept envisions that a synergistic action of these master regulator proteins at certain early-life and late-life checkpoints modulates the rates and efficiencies of progression of such processes as cell metabolism, growth, proliferation, stress resistance, macromolecular homeostasis, survival and death. The concept predicts that, by modulating these vital cellular processes throughout lifespan (i.e., prior to an arrest of cell growth and division, and following such arrest), the checkpoint-specific master regulator proteins orchestrate the development and maintenance of a pro- or anti-aging cellular pattern and, thus, define longevity of chronologically aging yeast.
INTRODUCTION
The budding yeast S. cerevisiae is an advantageous model organism for unveiling fundamental mechanisms and biological principles underlying the inherent complexity of cellular aging in multicellular eukaryotes [1][2][3][4][5][6]. Because this unicellular eukaryote is amenable to comprehensive biochemical, genetic, cell biological, chemical biological, system biological and microfluidic dissection analyses [7][8][9][10][11][12], its use as a model in aging research provided deep mechanistic insights into cellular processes essential for longevity regulation in evolutionarily distant eukaryotic organisms. Due to the relatively short and easily monitored chronological and replicative lifespans of the yeast S. cerevisiae, it played a pivotal role in discovering: (1) numerous genes that impact cellular aging and define organismal longevity not only in yeast but also in eukaryotic organisms across phyla; (2) some key nutrient- and energy-sensing signaling pathways that orchestrate an evolutionarily conserved set of longevity-defining cellular processes across species; and (3) several aging-decelerating and longevity-extending small molecules, many of which slow down aging, improve health, attenuate age-related pathologies and delay the onset of age-related diseases in evolutionarily distant multicellular eukaryotic organisms [1][2][3][4][5][6][13][14][15][16][17][18][19][20][21][22]. These studies convincingly demonstrated that the signaling pathways governing cellular aging and mechanisms of their modulation by longevity-extending genetic, dietary and pharmacological interventions are conserved across species.
–
There are two different paradigms of yeast aging. Each of them is traditionally investigated separately from each other with the help of robust assays. These assays are conducted under controllable laboratory conditions [23][24][25][26] and have been recently automated to enable a systems-level analysis of the aging process in a high-throughput format [9][10][11][12][27][28][29]. In the chronological aging paradigm, yeast aging is defined by the length of time during which a cell remains viable after an arrest of its growth and division [23][30][31]. Yeast chronological aging under laboratory conditions is assessed using a simple clonogenic assay. This assay measures the percentage of yeast cells that in liquid cultures remain viable at different time points following entry of a cell population into the non-proliferative stationary phase. Cell viability in the clonogenic assay is assessed by monitoring the ability of a cell to form a colony on the surface of a solid nutrient-rich medium [3][25][30]. Chronological aging in yeast is believed: (1) to mimic aging of non-dividing, post-mitotic cells (such as neurons) in a multicellular eukaryotic organism; and (2) to serve as a simple model for organismal aging [32][33]. In the replicative aging paradigm, yeast aging is defined by the maximum number of daughter cells that a mother cell can produce before becoming senescent [24][34][35]. S. cerevisiae reproduces by asymmetric cell division; therefore, its replicative aging under laboratory conditions is typically assessed by using a micromanipulator to remove the budding progeny of a mother cell and counting the cumulative number of asymmetric mitotic divisions this mother cell could undergo [24][35]. Replicative aging in yeast is thought to mimic aging of dividing, mitotically active cells (such as lymphocytes) in a multicellular eukaryotic organism [2][32]. The use of robust assays for elucidating longevity regulation in chronologically or replicatively aging yeast under controllable laboratory conditions has significantly advanced our understanding of cell-autonomous mechanisms that orchestrate longevity-defining cellular processes within an individual cell in eukaryotic organisms across phyla [1][2][3][5][6][10].
–
Recent studies in yeast also advanced fundamental knowledge about cell-non-autonomous intraspecies mechanisms of longevity regulation. Such mechanisms operate within organized populations of yeast cells that are attached to solid surfaces to form a colony or a biofilm; these cells: (1) communicate with each other and cells in surrounding colonies or biofilms; (2) age chronologically and replicatively; and (3) undergo spatially organized growth, differentiation, aging or death, depending on their position within the colony [4][36][37][38][39][40][41][42][43].
–
It seems that cell-autonomous and cell-non-autonomous intraspecies mechanisms regulating yeast longevity have evolved in the process of natural selection within an ecosystem [44][45][46]. It has been recently proposed that this process: (1) is governed by ecosystemic interspecies mechanisms of lifespan regulation operating within the ecosystem; and (2) is driven by the ability of yeast cells to undergo specific pro-survival changes in their metabolism and physiology in response to some chemical compounds that, after being released to the ecosystem by other groups of organisms, may trigger a hormetic and/or cytostatic response in yeast [44][45][46][47][48].
–
In this review we analyze recent progress in understanding of cell-autonomous mechanisms of chronological aging in the yeast S. cerevisiae. Our analysis suggests a concept of a biomolecular network underlying the chronology of cellular aging in yeast. This concept envisions that: (1) the network integrates the vital processes of cell metabolism, growth, proliferation, stress resistance, macromolecular homeostasis, survival and death; (2) the network progresses through a series of the early-life and late-life ʺcheckpointsʺ; (3) a gradual progression of the network through these lifespan checkpoints is monitored by some checkpoint-specific ʺmaster regulatorʺ proteins; and (4) such progression is critically important for establishing the pace of cellular aging.
CELL-AUTONOMOUS MECHANISMS ORCHESTRATE LONGEVITY-DEFINING CELLULAR PROCESSES IN CHRONOLOGICALLY AGING YEAST PRIOR TO AN ARREST OF CELL GROWTH AND DIVISION
A body of recent evidence supports the view that certain cellular processes taking place early in life of a chronologically aging yeast cell, prior to entry into a non-proliferative state, define the length of time during which this cell remains viable after such entry – i.e., define longevity of chronologically aging yeast grown under controllable laboratory conditions in liquid media [3][20][23][49][50][51][52][53][54][55][56][57][58][59][60][61][62][63][64][65][66][67][68][69][70][71][72][73]. These longevity-defining cellular processes: (1) are essential for metabolism, growth, proliferation, stress resistance, macromolecular homeostasis, survival and death of individual yeast cells that age chronologically; and (2) are orchestrated via cell-autonomous mechanisms of lifespan regulation operating within these cells. The longevity-defining cellular processes and mechanisms orchestrating their progression in chronologically aging yeast prior to entry into a non-proliferative state (and, for some of them, after such entry) are outlined below in this section.
–
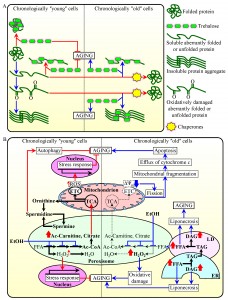
Intracellular trehalose modulates cellular protein homeostasis (proteostasis)
Recent studies revealed that the intracellular concentrations of trehalose prior to cell entry into a non-proliferative state and following such entry play essential and differing roles in defining longevity of chronologically aging yeast. This is because trehalose is involved in modulating protein folding, misfolding, unfolding, refolding, oxidative damage, solubility and aggregation throughout lifespan [23][61] (Figure 1A). In chronologically ʺyoungʺ yeast cells, which undergo growth and division, trehalose plays an essential longevity-extending role because this non-reducing disaccharide: (1) binds to newly synthesized cellular proteins, thereby stabilizing their native folding states and attenuating their conversion into aberrantly folded and/or unfolded protein species; (2) shields the contiguous exposed hydrophobic side chains of amino acids that are abundant in misfolded, partially folded and unfolded protein species and that are known to promote their aggregation, thereby eliciting a direct inhibitory effect on the formation of insoluble protein aggregates; and (3) protects cellular proteins from oxidative carbonylation by interacting with their carbonylation-prone aberrantly folded species, thus having an indirect inhibitory effect on the aggregation of oxidatively damaged proteins [23][61] (Figure 1A).
In contrast, in chronologically ʺoldʺ yeast cells, which do not grow or divide, trehalose plays a key role in shortening longevity. This is because in such cells trehalose shields the patches of hydrophobic amino acid residues that are abundant in aberrantly folded protein species [61]. Thus, in chronologically ʺoldʺ yeast cells trehalose competes with molecular chaperones for binding with these patches of hydrophobic amino acid residues known to be required for the chaperone-assisted refolding of misfolded, partially folded and unfolded protein species [74][75][76][77] – either soluble or extracted from protein aggregates with the help of molecular chaperones [61] (Figure 1A). Importantly, it has been demonstrated that a caloric restriction (CR) diet and certain genetic interventions affecting trehalose synthesis or degradation extend longevity of chronologically aging yeast because they simultaneously: (1) increase the intracellular concentration of trehalose by 70-160% (above its level detected in yeast cultured under non-CR conditions) in chronologically ʺyoungʺ, proliferating cells; and (2) reduce the intracellular concentration of trehalose by 60-80% (below a threshold observed in yeast cultured under non-CR conditions) in chronologically ʺoldʺ, non-proliferating cells [61]. These findings suggest the existence of at least two ʺcheckpointsʺ during the lifespan of a chronologically aging yeast cell at which the intracellular concentration of trehalose (which depends on a balance between trehalose synthesis and degradation) defines its longevity by modulating cellular proteostasis. It seems that one of these checkpoints exists early in life of a chronologically aging yeast cell (i.e., prior to entry into a non-proliferative state), whereas the other checkpoint occurs late in its life (i.e., after such cell enters a non-proliferative state).
–
Protein import into the peroxisome impacts longevity-defining processes in other cellular compartments
The efficiency of peroxisomal protein import has been shown to decline with the chronological age of a eukaryotic cell [57][58][78][79]. Such import is driven by Pex5p and Pex7p, the peroxisomal targeting signal type 1 (PTS1) and PTS2 cytosolic shuttling receptors, respectively [80][81][82]. Recent findings support the view that the age-dependent efficiency of protein import into the peroxisome defines the efficiencies of fatty acid oxidation, hydrogen peroxide turnover and anaplerotic metabolism within this organelle. These metabolic processes are known to modulate the dynamic communications of peroxisomes with other cellular compartments via a unidirectional or bidirectional flow of certain soluble metabolites and lipids [20][57][58][65][83][84][85][86][87]. By influencing longevity-defining cellular processes confined to these other compartments, the metabolic processes within the peroxisome cause the development of a pro- or anti-aging cellular pattern [20][54][57][58][65][88][89][83][84][85][86][87].
–
Altogether, these findings suggest a model for how the age-dependent efficiency of peroxisomal protein import in chronologically aging yeast defines the age-related metabolic pattern of peroxisomes, thus impacting longevity-defining processes in other cellular compartments and ultimately establishing a pro- or anti-aging cellular pattern (Figure 1B). The model envisions that chronologically ʺyoungʺ yeast cells develop and maintain an anti-aging cellular pattern in part because in these cells the efficiency of Pex5p- and Pex7p-dependent peroxisomal protein import exceeds a threshold. Specifically, such ʺyoungʺ cells are proficient in peroxisomal import of the following proteins: (1) catalase Cta1p and peroxiredoxin Pmp20p, both required for decomposition of hydrogen peroxide and other reactive oxygen species (ROS) within the peroxisome; (2) Fox1p, Fox2p and Fox3p, enzymes involved in peroxisomal β-oxidation of fatty acids to acetyl-CoA; and (3) the citrate synthase Cit2p and acetyl-carnitine synthase Cat2p, both facilitating the replenishment of tricarboxylic acid (TCA) cycle intermediates destined for mitochondria by catalyzing the anaplerotic conversion of acetyl-CoA to citrate and acetyl-carnitine, respectively [14][20][57][58][83][90][91][92] (Figure 1B). The efficient peroxisomal import of all these proteins in chronologically ʺyoungʺ yeast cells enables the establishment of an anti-aging cellular pattern by: (1) minimizing the oxidative damage to peroxisomal proteins and membrane lipids; (2) maintaining the intracellular concentration of peroxisomally produced hydrogen peroxide at a threshold which is insufficient to damage cellular macromolecules but can activate transcription of nuclear genes essential for cell survival, thus promoting the longevity-extending cellular process of “stress-response hormesis”; (3) stimulating the TCA cycle and electron transport chain (ETC) in mitochondria, thus enabling to sustain mitochondrially generated ROS at a non-toxic level which is sufficient to stimulate transcription of nuclear genes encoding stress-protecting and other anti-aging proteins [20][23][57][58][65][92] (Figure 1B).
–
Noteworthy, peroxisomes in yeast house the polyamine oxidase Fms1p, an enzyme involved in the synthesis of spermidine [17][93]. This natural polyamine has been shown to extend longevity of chronologically aging yeast by stimulating the essential cytoprotective cellular process of autophagy [13][94][95] (Figure 1B). Because the intracellular concentration of spermidine in chronologically ʺyoungʺ yeast exceeds that in chronologically ʺoldʺ yeast [13], one could speculate that peroxisomal import of Fms1p early in life of a chronologically aging yeast cell is more efficient than it is late in life, after entry of a chronologically aging yeast cell population into a non-proliferative state. In this scenario, chronologically ʺyoungʺ yeast cells develop and maintain an anti-aging cellular pattern in part because they are proficient in peroxisomal import of a protein needed for the synthesis of a natural polyamine which promotes the longevity-extending cellular process of autophagy (Figure 1B).
–
Our model further posits that in chronologically ʺoldʺ yeast cells the efficiencies of Pex5p- and Pex7p-dependent peroxisomal import of Cta1p, Pmp20p, Fox1p, Fox2p, Fox3p, Cit2p and Cat2p markedly decline (Figure 1B). Such deterioration of peroxisomal protein import below a threshold in these cells causes the development of a pro-aging cellular pattern by: (1) elevating the intracellular concentration of peroxisomally produced hydrogen peroxide above a cytotoxic level, thus increasing the extent of oxidative damage to cellular macromolecules; (2) reducing the efficiency of Fox1p-, Fox2p- and Fox3p-driven peroxisomal oxidation of fatty acids derived from triacylglycerols that are synthesized in the endoplasmic reticulum (ER) and deposited within lipid droplets (LD) – thus elevating the concentrations of non-esterified (ʺfreeʺ) fatty acids and diacylglycerol, both of which are known to elicit an age-related form of liponecrotic programmed cell death (PCD); (3) diminishing the replenishment of TCA cycle intermediates destined for the TCA cycle in mitochondria – thus triggering a cascade of events that reduce the ETC in mitochondria, lower electrochemical potential across the inner mitochondrial membrane (IMM), promote mitochondrial fragmentation, cause the efflux of cytochrome c and other pro-apoptotic proteins from fragmented mitochondria, and ultimately elicit an age-related form of apoptotic PCD [14][20][23][54][57][58][65][89][91][92][96][97][98][99][100][101][102][103] (Figure 1B).
–
Furthermore, our model envisions that chronologically ʺoldʺ yeast cells develop and maintain a pro-aging cellular pattern in part because they exhibit a low efficiency of peroxisomal import of the polyamine oxidase Fms1p, which is required for the synthesis of spermidine [13][17][93][94][95] (Figure 1B).
–
Noteworthy, both chronologically ʺyoungʺ and ʺoldʺ yeast cells grown in a nutrient-rich medium under longevity-shortening non-CR conditions have been shown to accumulate ethanol, a product of glucose fermentation [23][89]. Ethanol is known to repress the synthesis of Fox1p, Fox2p and Fox3p, thereby suppressing Fox1p-, Fox2p- and Fox3p-driven peroxisomal oxidation of fatty acids [90][104] (Figure 1B). By suppressing peroxisomal oxidation of fatty acids to acetyl-CoA in chronologically ʺyoungʺ cells under non-CR conditions, the accumulated ethanol attenuates the anaplerotic conversion of acetyl-CoA to citrate and acetyl-carnitine – thus inhibiting an aging-decelerating cellular process of the replenishment of TCA cycle intermediates destined for mitochondria [57][58][90][104] (Figure 1B). Moreover, by suppressing peroxisomal oxidation of fatty acids to acetyl-CoA in chronologically ʺoldʺ cells under non-CR conditions, the accumulated ethanol also elevates the concentrations of non-esterified (ʺfreeʺ) fatty acids and diacylglycerol – thus triggering an age-related form of liponecrotic PCD (Figure 1B) [14][23][54][57][58][89][103]. It should be stressed that neither chronologically ʺyoungʺ nor chronologically ʺoldʺ yeast cells grown under longevity-extending CR conditions amass ethanol [23][89]. Such inability of yeast cells limited in calorie supply to accumulate ethanol is one of the reasons of why they are able to develop and maintain an anti-aging cellular pattern throughout lifespan [23][54][57][58][89] (Figure 1B).
–
In sum, it is conceivable that there are at least two checkpoints during the lifespan of a chronologically aging yeast cell at which the age-dependent efficiency of peroxisomal protein import defines the age-related metabolic pattern of peroxisomes, thus impacting longevity-defining processes in other cellular compartments and ultimately establishing a pro- or anti-aging cellular pattern. It seems that one of these checkpoints occurs early in life of a chronologically aging yeast cell (i.e., prior to an arrest of cell growth and division), whereas the other checkpoint exists late in its life (i.e., following such arrest).
–
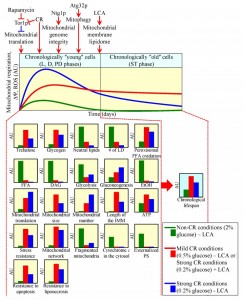
Coupled mitochondrial respiration, mitochondrial membrane potential and mitochondrial ROS production affect longevity-defining processes in other cellular locations
The functional state of mitochondria and mitochondrial ROS production early in life of a chronologically aging yeast cell, prior to entry into a non-proliferative state, have been shown to define the length of time during which this cell remains viable after such entry – i.e., define longevity of chronologically aging yeast [3][14][23][31][49][50][51][53][55][56][62][64][68][69][70][71][72][73][105][106][107][108]. One key feature of the longevity-defining functional state of mitochondria in chronologically ʺyoungʺ, proliferating yeast cells is the capacity of electron transport along the respiratory chain coupled to ATP synthesis [3][14][23][49][50][51][62][64][70][72][73][106][107]. Another such key feature is the value of mitochondrial membrane potential; it depends on a balance between the capacities of ETC-driven proton transport from the matrix to the intermembrane space and proton translocation across the IMM in the opposite direction [3][14][23][50][51][56][64][70]. Moreover, the longevity-defining process of mitochondrial ROS production in chronologically ʺyoungʺ yeast cells depends on the efficiency of coupling between the ETC and oxidative phosphorylation (OXPHOS) system in mitochondria [50][51].
–
Recent studies revealed how various genetic, dietary and pharmacological interventions having diverse effects on the ETC, OXPHOS system and/or ROS production in mitochondria of chronologically ʺyoungʺ yeast impact yeast longevity [3][14][23][31][49][50][51][53][55][56][62][64][68][69][70][71][72][73][105][106][107][108]. These studies suggest a model for how coupled mitochondrial respiration, mitochondrial membrane potential maintenance and mitochondrial ROS production early in life of chronologically aging yeast cells define their longevity. This model is depicted schematically in Figure 2. The model envisions that chronologically ʺyoungʺ yeast cells cultured under non-CR conditions develop and maintain a pro-aging cellular pattern because the capacities of these mitochondrial processes in such cells are below a certain level (Figure 2; these capacities are displayed in green color) [23][53][62][72][73]. Furthermore, the model posits that chronologically ʺyoungʺ yeast cells, in which the capacities of these mitochondrial processes exceed a critical threshold, develop and maintain an anti-aging cellular pattern. This is because such capacities have specific impacts on: (1) some longevity-defining processes confined to mitochondria; and (2) certain longevity-defining processes in other cellular locations (Figure 2; these capacities are displayed in red color) [14][23][50][51][56][62][64][70][73][107]. The model also predicts that a significant further increase in the capacities of coupled mitochondrial respiration, mitochondrial membrane potential maintenance and mitochondrial ROS production above a critical threshold in chronologically ʺyoungʺ yeast cells has a negative impact on their longevity (Figure 2; these capacities are displayed in blue color) [14][23][64][107].
The capacities of coupled mitochondrial respiration, mitochondrial membrane potential maintenance and mitochondrial ROS production in chronologically ʺyoungʺ yeast cells are modulated by several ʺupstreamʺ pathways (Figure 2). These longevity-defining pathways include: (1) the nutrient- and energy-sensing TOR (target of rapamycin) signaling pathway, which through the rapamycin-sensitive protein kinase Tor1p inhibits mitochondrial translation of the OXPHOS enzymes encoded by mitochondrial DNA (mtDNA) [14][50][51][56][62]; (2) a CR pathway, whose ability to modulate coupled mitochondrial respiration, mitochondrial membrane potential maintenance and mitochondrial ROS production in chronologically ʺyoungʺ yeast cells is mediated in part by Tor1p [3][14][23][33][50][51][52][56][62]; (3) a pathway for the maintenance of mitochondrial genome integrity and copy number, which is orchestrated by the mitochondrial base-excision repair enzyme Ntg1p [108]; (4) the mitophagy pathway of mitochondrial quality control responsible for autophagic degradation of aged, dysfunctional or damaged mitochondria – which requires the receptor protein Atg32p on the surface of mitochondria destined for such degradation [70]; and (5) a pathway for specific remodeling of the membrane lipidome of mitochondria – which is stimulated in response to accumulation of the exogenously added lithocholic acid (LCA), an anti-aging natural compound, predominantly in the IMM [64][107] (Figure 2). Noteworthy, some of these ʺupstreamʺ pathways overlap; such convergent pathways modulating the capacities of coupled mitochondrial respiration, mitochondrial membrane potential maintenance and mitochondrial ROS production in chronologically ʺyoungʺ yeast include the TOR and CR pathways [3][14][23][33][50][51][52][56][62], as well as the TOR and Ntg1p-governed pathways [108]. In contrast, it seems that other ʺupstreamʺ pathways modulating the capacities of these three longevity-defining mitochondrial processes do not converge and act in synergy; such ʺparallelʺ pathways include: (1) a CR pathway and an LCA-driven pathway for remodeling of the membrane lipidome of mitochondria [64][107]; and (2) a CR pathway and the Ntg1p-governed pathway [108].
–
The capacities of coupled mitochondrial respiration, mitochondrial membrane potential maintenance and mitochondrial ROS production in chronologically ʺyoungʺ yeast cells define their longevity by orchestrating numerous ʺdownstreamʺ cellular processes throughout lifespan – i.e., prior to entry into a non-proliferative state and after such entry (Figure 2). Among them are the following ʺdownstreamʺ processes (Figure 2): (1) the maintenance of trehalose homeostasis, a longevity-defining process known to modulate proteostasis in chronologically ʺyoungʺ and ʺoldʺ cells [23][61] (Figure 1A); (2) the maintenance of the homeostasis of glycogen, a reserve carbohydrate whose elevated level in chronologically ʺyoungʺ and ʺoldʺ cells is a hallmark of carbohydrate metabolism remodeling in yeast cultured under CR [23]; (3) the maintenance of the homeostasis of neutral lipids deposited within LD, a process known to play an essential role in regulating longevity of chronologically aging yeast [14][23][92] (Figure 1B); (4) peroxisomal oxidation of fatty acids, a process implicated in yeast chronological aging [14][23][91][92] (Figure 1B); (5) the maintenance of the homeostasis of non-esterified (ʺfreeʺ) fatty acids and diacylglycerol, whose reduced levels in chronologically ʺyoungʺ and ʺoldʺ cells are characteristic of lipid metabolism remodeling in yeast cultured under CR – a pattern likely linked to the demonstrated abilities of both lipid species to elicit an age-related form of liponecrotic PCD [14][23][103] (Figure 1B); (6) the maintenance of a balance between the relative rates of glycolysis and gluconeogenesis, a process known to impact the level of ethanol in chronologically ʺyoungʺ and ʺoldʺ yeast – thus defining the extent to which this product of glucose fermentation suppresses the longevity-extending process of peroxisomal oxidation of fatty acids [14][23] (Figure 1B); (7) the longevity-extending process of mitochondrial translation [23][50][51][56]; (8) the maintenance of a balance between the relative rates of mitochondrial fusion and fission, a process known to define the size and number of mitochondria, the length of mitochondrial cristae extending from the IMM, and the level of ATP synthesized in mitochondria of chronologically ʺyoungʺ and ʺoldʺ yeast [14][23][64][70]; (9) the development of an age-related pattern of susceptibility to chronic oxidative, thermal and osmotic stresses [3][14][23][33][50][56][59][62]; (10) an age-related form of apoptotic PCD, which in chronologically ʺyoungʺ yeast is manifested in such early hallmark events of this PCD as the fragmentation of a tubular mitochondrial network into individual mitochondria, release of cytochrome c from mitochondria into the cytosol and phosphatidylserine (PS) translocation from the inner to the outer leaflet of the plasma membrane [14][23] (Figure 1B); and (11) the development of a pattern of cell susceptibility to an age-related forms of apoptotic and liponecrotic PCD elicited by an exposure to exogenous hydrogen peroxide or palmitoleic acid, respectively [14][23][59][103].
–
The molecular mechanisms through which the capacity of mitochondrial ROS production in chronologically ʺyoungʺ yeast cells defines their longevity have begun to emerge; they involve communication between mitochondria and the nucleus via two signaling pathways [71][108]. In one of these signaling pathways, hormetic concentrations of ROS released from mitochondria trigger a pro-longevity transcriptional program in the nucleus by stimulating Gis1p, Msn2p and Msn4p [68][71][108]; these transcriptional factors are known to activate expression of numerous genes essential for the resistance to various stresses, stationary phase survival, carbohydrate metabolism, nutrient sensing and chronological longevity assurance [109][110][111]. Another mitochondria-to-nucleus signaling pathway initiated by hormetic concentrations of ROS in chronologically ʺyoungʺ yeast cells involves a cascade of events within the nucleus. In this cascade, the DNA damage response (DDR) kinase Tel1p responds to hormetic concentrations of ROS released from mitochondria by phosphorylating and activating the DDR kinase Rad53p. Active Rad53p then phosphorylates and inactivates the histone demethylase Rph1p confined to subtelomeric chromatin regions, thereby repressing their transcription, minimizing telomeric DNA damage and ultimately extending longevity of chronologically aging yeast [68][71]. Noteworthy, it seems that the Tel1p-Rad53p-Rph1p signaling pathway overlaps with and is regulated by the ʺupstreamʺ Ntg1p-governed pathway (Figure 2) for the maintenance of mitochondrial genome integrity and copy number [108].
–
Altogether, these findings suggest that there is a checkpoint early in life of chronologically aging yeast cells (i.e., prior to entry into a non-proliferative state) at which the capacities of coupled mitochondrial respiration, mitochondrial membrane potential maintenance and mitochondrial ROS production define their viability after such entry – i.e., define their longevity. It seems that the capacities of these three mitochondrial processes at such checkpoint define yeast longevity by orchestrating a number of ʺdownstreamʺ processes taking place in various cellular locations throughout lifespan, before an arrest of cell growth and division, and following such arrest.
–
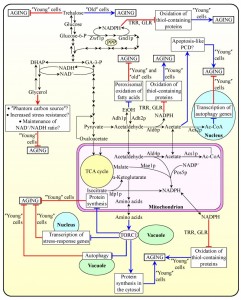
Metabolite flow within glycolytic and non-glycolytic pathways of carbohydrate metabolism defines the establishment of a pro- or anti-aging cellular pattern
Recent studies provided evidence that the relative rates of reactions comprising glycolytic and non-glycolytic pathways of carbohydrate metabolism, as well as the intracellular concentrations of some key intermediates in these pathways, define the development and maintenance of a pro- or anti-aging cellular pattern in chronologically aging yeast [3][10][23][33][52][54][57][58][63][65][66][67][69][89][112][113][114][115][116][117]. The major findings of these studies can be summarized as follows: (1) the establishment of a certain metabolic pattern of such coordinated pathways early in life of chronologically aging yeast cells, prior to entry into a non-proliferative state, defines their longevity; and (2) some dietary, genetic and pharmacological interventions extend yeast longevity by causing a specific remodeling of such metabolic pathways in chronologically ʺyoungʺ, proliferating cells [23][52][66][67][69][112][113][114][115][116][117]. These findings suggest a model for how metabolic flux within the network integrating glycolytic and non-glycolytic pathways of carbohydrate metabolism modulates longevity-defining cellular processes. This model is depicted schematically in Figure 3.
The model posits that glucose, the primary carbon source used in most assays for investigating yeast chronological aging under laboratory conditions, is initially converted to pyruvate via the glycolytic pathway and also enters the pentose phosphate pathway (PPP) [23][25][27][30][33][57][66] (Figure 3). In chronologically ʺyoungʺ yeast cells progressing through logarithmic (L) phase, the PPP generates not only ribose-5-phosphate for nucleic acid synthesis but also NADPH, the primary source of cellular reducing equivalents required for the reductive synthesis of fatty acids, sterols and some amino acids [60][63][66][118]. Importantly, NADPH – which is produced in the Zwf1p- and Gnd1p-driven oxidative reactions of the PPP – also functions as the electron donor essential for sustaining cellular redox homeostasis via thioredoxin and glutathione reductase systems [63][66][118] (Figure 3). These two NADPH-dependent systems have been shown to reduce the extent of oxidative damage to numerous thiol-containing cytosolic, nuclear and mitochondrial proteins in chronologically ʺyoungʺ yeast cells; as such, both reductase systems play essential roles in longevity assurance and underlie, in part, the robust longevity-extending effect of CR [63][66] (Figure 3).
–
After chronologically aging yeast cells consume glucose during L phase, they enter diauxic (D) and then post-diauxic (PD) phases of slow growth [23][25][30][33]. During D and PD phases, prior to an arrest of cell growth and division and entry into the non-proliferative stationary (ST) phase, the pyruvate formed by glycolysis can enter several alternative pathways of carbon metabolism; all these pathways have been implicated in modulating various longevity-defining cellular processes [3][10][23][33][52][54][57][58][63][65][66][67][69][89][112][113][114][115][116][117]. One of these alternative metabolic pathways is fermentation leading to the formation of ethanol and/or acetic acid in the cytosol of a chronologically ʺyoungʺ yeast cell; the nature of a product of such fermentation depends on the type of a synthetic or nutrient-rich growth medium used and/or aeration conditions applied [3][10][23][33][52][112][113][114][115][117] (Figure 3). As discussed above, the level of ethanol in chronologically aging yeast defines the extent to which it suppresses the longevity-extending process of peroxisomal oxidation of fatty acids [14][23] (Figure 1B). The steady-state level of this product of glucose fermentation depends on the relative enzymatic activities of Adh1p and Adh2p, which are required for ethanol formation or oxidation, respectively [3][23][52][112][117] (Figure 3). Acetic acid, the alternative product of glucose fermentation in the cytosol of chronologically ʺyoungʺ yeast cells, shortens their longevity – likely because it can elicit an age-related form of apoptotic PCD [3][27][113][114][115] (Figure 3). Of note, the Ald6p-dependent acetaldehyde dehydrogenase reaction in the cytosol of these cells yields not only acetic acid but also NADPH [60][66][118] (Figure 3). As discussed above, the NADPH-dependent thioredoxin and glutathione reductase systems are vital for longevity assurance and are essential for the longevity-extending effect of CR because they reduce the extent of oxidative damage to many thiol-containing cellular proteins in chronologically ʺyoungʺ yeast cells [63][66][118] (Figure 3). Moreover, acetic acid can be converted to acetyl-CoA in the nucleo-cytosolic Acs2p-dependent reaction [116][118]. The acetyl-CoA formed in the nucleus has been shown to shorten longevity of chronologically aging yeast by causing histone H3 hyperacetylation, thereby selectively suppressing transcription of the ATG5, ATG7, ATG11 and ATG14 genes; these genes encode proteins needed for the longevity-extending process of autophagy [116] (Figure 3). It remains to be determine what are the relative impacts of acetic acid (a pro-aging metabolite), NADPH (an anti-aging metabolite) and acetyl-CoA (a pro-aging metabolite) on longevity of chronologically aging yeast.
–
The model depicted in Figure 3 further envisions that glucose fermentation to glycerol in the cytosol of a chronologically ʺyoungʺ yeast cell operates as a longevity-extending cellular process – likely because it reduces metabolite flow into the longevity-shortening cellular process of glucose fermentation to ethanol and/or acetic acid [52][113] (Figure 3). The term ʺphantom carbon sourceʺ has been coined for defining this aspect of the essential longevity-extending role of glycerol in yeast [52]. Glycerol formed by glucose fermentation in the cytosol of a chronologically ʺyoungʺ yeast cell has been proposed to extend it lifespan also because it is known to reduce cell susceptibility to chronic oxidative, thermal and osmotic stresses [52] (Figure 3). Furthermore, glucose fermentation to glycerol in chronologically ʺyoungʺ yeast cells may also play an essential role in longevity assurance by maintaining an NAD+/NADH ratio characteristic of an anti-aging cellular pattern [52] (Figure 3).
–
In addition to being converted to ethanol, acetic acid, NADPH and/or acetyl-CoA, the pyruvate formed by glycolysis can enter the gluconeogenesis pathway leading to the formation of glucose; in chronologically aging yeast, this newly synthesized glucose can be further used for the synthesis of trehalose [23][61][62][117] (Figure 3). As discussed above [23][61] (Figure 1A): (1) in chronologically ʺyoungʺ yeast cells progressing through D and PD growth phases, trehalose plays an essential longevity-extending role, whereas (2) in chronologically ʺoldʺ, non-proliferating cells, trehalose plays a key role in shortening longevity (Figure 1).
–
In chronologically ʺyoungʺ yeast cells progressing through D and PD growth phases, the pyruvate formed by glycolysis – as well as the acetaldehyde and acetate derived from it – can also fuel several longevity-defining metabolic processes in mitochondria. One of these metabolic processes is the TCA cycle. Two intermediates of the cycle, malate and isocitrate, can be used to form NADPH in the Mae1p- and Idp1p-dependent reactions, respectively [60][66][118] (Figure 3). NADPH in mitochondria of chronologically ʺyoungʺ yeast cells progressing through D and PD growth phases can also be formed in the Ald4p-dependent acetaldehyde dehydrogenase reaction and in the Pos5p-dependent NADH kinase reaction [60][66][118] (Figure 3). As discussed above, NADPH can play a vital longevity-extending role by being used for a thioredoxin- and glutathione reductase-driven decrease in the extent of oxidative damage to thiol-containing mitochondrial proteins and proteins in other cellular locations [63][66][118] (Figure 3). Moreover, the oxaloacetate and α-ketoglutarate intermediates of the TCA cycle in mitochondria of chronologically ʺyoungʺ yeast cells progressing through D and PD growth phases are known to be used for the synthesis of amino acids [60][118][119] (Figure 3). After their exit from mitochondria to the cytosol, some of these amino acids – including aspartate, asparagine, glutamate and glutamine – cause a significant increase in protein kinase activity of the TOR complex 1 (TORC1) on the surface of vacuoles [20][119][120][121][122][123][124] (Figure 3). The resulting stimulation of the TOR signaling pathway in chronologically ʺyoungʺ yeast cells is known to initiate the establishment of a pro-aging cellular pattern by: (1) activating the longevity-shortening process of protein synthesis in the cytosol; (2) suppressing the longevity-extending process of autophagy in vacuoles; (3) inhibiting the longevity-extending process of transcription of numerous stress-response genes in the nucleus; and (4) suppressing the longevity-extending process of protein synthesis in mitochondria [20][50][51][119][120][121][122][123][124] (Figure 3).
–
Taken together, these findings strongly suggest that there are several checkpoints early in life of chronologically aging yeast cells – during L, D and PD phases preceding entry into the non-proliferative ST phase – at which the coordinated metabolite flow within glycolytic and non-glycolytic pathways of carbohydrate metabolism defines yeast longevity. It seems that at each of these early-life checkpoints some key intermediates in such pathways affect – in a different manner and in a concentration-dependent fashion – the vital processes of cell metabolism, growth, proliferation, stress resistance, macromolecular homeostasis, survival and death. By modulating such longevity-defining cellular processes throughout lifespan – prior to an arrest of cell growth and division and following such arrest – these key metabolic intermediates define the development and maintenance of a pro- or anti-aging cellular pattern.
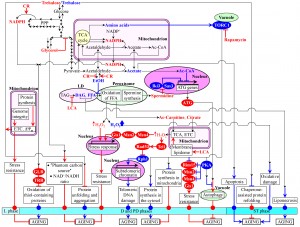
A STEPWISE PROGRESSION OF A BIOMOLECULAR NETWORK OF CELLULAR AGING THROUGH A SERIES OF LIFESPAN CHECKPOINTS DEFINES LONGEVITY OF CHRONOLOGICALLY AGING YEAST
The above analysis of the current knowledge about cell-autonomous mechanisms underlying chronological aging in yeast suggests the existence of several lifespan checkpoints that are critically important for establishing the pace of such aging. A recently reported ability of a natural chemical compound to extend longevity of chronologically aging yeast only if added at some of these checkpoints [59] supports the notion that a stepwise progression through such checkpoints may define the development and maintenance of a pro- or anti-aging cellular pattern. Therefore, in this review we extend the recently proposed network theories of yeast chronological aging [3][20][23][58][59][61][62][125][126][127] by putting forward a concept of a biomolecular network underlying the chronology of cellular aging in yeast. This concept is depicted schematically in Figure 4. The concept posits that the network progresses through a series of the early-life checkpoints (that exist in L, D and PD phases) and late-life checkpoints (that exist in ST phase). At each of these checkpoints, the intracellular concentrations of some key intermediates and products of certain metabolic pathways – as well as the rates of coordinated flow of such metabolites within an intricate network of intercompartmental (i.e., organelle-organelle and organelle-cytosol) communications – are monitored by some checkpoint-specific ʺmaster regulatorʺ proteins (Figure 4). The concept further envisions that, because each of these master regulator proteins is known for its essential role in longevity regulation, their synergistic action at certain early-life and late-life checkpoints modulates the rates and efficiencies of progression of such essential processes as cell metabolism, growth, proliferation, stress resistance, macromolecular homeostasis, survival and death (Figure 4). The concept predicts that, by modulating these vital cellular processes throughout lifespan – prior to an arrest of cell growth and division and following such arrest – the checkpoint-specific master regulator proteins orchestrate the development and maintenance of a pro- or anti-aging cellular pattern and, thus, define longevity of chronologically aging yeast (Figure 4).
In the proposed concept of a biomolecular network underlying the chronology of cellular aging in yeast, the key intermediates and products of longevity-defining metabolic pathways include the following metabolites: NADPH (an anti-aging metabolite), glycerol (an anti-aging metabolite), trehalose (an anti-aging metabolite in ʺyoungʺ cells but a pro-aging metabolite in ʺoldʺ cells), acetyl-carnitine (an anti-aging metabolite), citrate (an anti-aging metabolite), hydrogen peroxide (an anti-aging metabolite if it is maintained at an ʺadaptiveʺ level eliciting a hormetic response but a pro-aging metabolite if it amasses above a cytotoxic level), spermidine (an anti-aging metabolite), ethanol (a pro-aging metabolite), acetic acid (a pro-aging metabolite), acetyl-CoA (a pro-aging metabolite), non-esterified fatty acids (pro-aging metabolites), diacylglycerol (a pro-aging metabolite) and amino acids (pro-aging metabolites) (Figure 4). The proposed concept posits that these key metabolites undergo an age-related flow within an intricate network of intercompartmental communications. Such unidirectional and bidirectional flow of the critical metabolites between different cellular compartments connects mitochondria and the nucleus, peroxisomes and the nucleus, vacuoles and the nucleus, the cytosol and the nucleus, mitochondria and peroxisomes, lipid droplets and peroxisomes, mitochondria and vacuoles, peroxisomes and the cytosol, mitochondria and the cytosol, and vacuoles and the cytosol (Figure 4). In the proposed concept, a CR diet and some pharmacological interventions (such as rapamycin and LCA) – as well as some other environmental cues (such as the intake of certain dietary supplements and hormetic environmental stresses) – extend longevity of chronologically aging yeast by altering the age-related dynamics of changes in the intracellular concentrations of the key metabolites and/or by modulating the intercompartmental flow of these critical metabolites (Figure 4). The following proteins and protein complexes operate as checkpoint-specific master regulators that – according to the proposed concept – respond to age-related changes in the intracellular concentrations of the key metabolites and in the intensity of their intercompartmental flow by modulating the longevity-defining processes of cell metabolism, growth, proliferation, stress resistance, macromolecular homeostasis, survival and death: (1) thioredoxin and glutathione reductase systems (anti-aging master regulators); (2) Rim15p, Msn2p, Msn4p and Gis1p (anti-aging master regulators); (3) Tel1p and Rad53p (anti-aging master regulators); (4) components of the ATG protein machinery involved in the essential cytoprotective cellular process of autophagy (anti-aging master regulators); (5) Rph1p (a pro-aging master regulator); (6) Iki3p and Sas3p (pro-aging master regulators); and (7) TORC1 and protein kinase A (PKA) (pro-aging master regulators) (Figure 4). Future studies are likely to reveal some other checkpoint-specific master regulator proteins. Finally, the proposed concept posits that the checkpoint-specific master regulators define longevity of chronologically aging yeast by orchestrating certain ʺdownstreamʺ cellular processes and features throughout lifespan – i.e., prior to entry into a non-proliferative state and after such entry (Figure 4). These ʺdownstreamʺ processes and features include (Figure 4): (1) susceptibility to chronic oxidative, thermal and osmotic stresses; (2) oxidative damage to numerous thiol-containing cytosolic, nuclear and mitochondrial proteins; (3) cellular proteostasis, which depends on the relative rates and efficiencies of protein folding, misfolding, unfolding, refolding, oxidative damage, solubility and aggregation; (4) transcription of subtelomeric chromatin regions, which defines the extent of age-related telomeric DNA damage; (5) protein synthesis in the cytosol; (6) protein synthesis in mitochondria; (7) autophagy, an essential mechanism of cellular quality control responsible for the autophagic degradation of aged, dysfunctional and damaged macromolecules and organelles; and (8) liponecrotic and apoptotic forms of age-related PCD.
CONCLUSIONS
Emergent evidence supports the view that the processes of cell metabolism, growth, proliferation, stress resistance, macromolecular homeostasis, survival and death in chronologically aging yeast are integrated into a biomolecular network of cellular aging. Recent findings imply that a stepwise progression of this network through a series of the early-life and late-life checkpoints is monitored by some checkpoint-specific master regulator proteins; these proteins act in synergy to orchestrate the development and maintenance of a pro- or anti-aging cellular pattern and, thus, to define longevity of chronologically aging yeast. The major challenge now is to understand if a process of yeast chronological aging – whose progression through a series of lifespan checkpoints is monitored and controlled by a distinct set of master regulator proteins – is a program. We hypothesize that, although yeast chronological aging is a gradual and controllable process progressing through a series of consecutive lifespan checkpoints, it is not a program aimed at the termination of lives of individual yeast cells and of their organized populations. Rather, chronological aging in yeast is probably due to the inability of chronologically ʺyoungʺ, proliferating cells to maintain the capacities of some crucial cellular processes above a critical threshold; these key cellular processes may include ones that ensure robust cell growth and proliferation, implement a spatiotemporal control of cell development and differentiation into quiescent and non-quiescent cell populations, and limit an age-related accumulation of molecular and cellular damage. The proposed here hypothesis also posits that the extreme cellular stress caused by the excessive accumulation of such damage in chronologically ʺoldʺ, non-proliferating cells may activate pathways known to orchestrate the apoptotic, regulated necrotic, autophagic and/or liponecrotic subroutines of PCD; these subroutines are believed to constitute modules that are dynamically integrated into a so-called PCD network [103][128][129][130][131][132][133][134][135]. In sum, the hypothesis of non-programmed chronological aging in yeast envisions that: (1) the processes of cell metabolism, growth, proliferation, stress response, macromolecular homeostasis, development and differentiation have evolved in the course of natural selection within diverse ecosystems (and perhaps under laboratory conditions) as programs aimed at sustaining the long-term survival of individual yeast cells under various environmental conditions; (2) the processes of various subroutines of PCD have evolved throughout natural selection within different ecosystems (and perhaps under laboratory conditions) as ʺaltruisticʺ programs aimed at sustaining the long-term survival of organized yeast populations (such as colonies and biofilms of yeast cells attached to solid surfaces) by eliminating individual yeast cells that are damaged, unable to mate and reproduce, poorly adapted to diverse environmental conditions, and/or release excessive quantities of ROS and other harmful metabolites (for a comprehensive discussion of this topic, see refs. [136][137]); and (3) a trade-off between these programs aimed at sustaining the long-term survival of individual yeast cells or that of organized yeast populations under diverse environmental conditions drives the evolution of yeast longevity towards maintaining a finite yeast chronological lifespan within an ecosystem. The proposed here hypothesis of non-programmed chronological aging in yeast provides a framework for future studies aimed at testing its validity.
References
- L. Fontana, L. Partridge, and V.D. Longo, "Extending Healthy Life Span—From Yeast to Humans", Science, vol. 328, pp. 321-326, 2010. http://dx.doi.org/10.1126/science.1172539
- M. Kaeberlein, "Lessons on longevity from budding yeast", Nature, vol. 464, pp. 513-519, 2010. http://dx.doi.org/10.1038/nature08981
- V. Longo, G. Shadel, M. Kaeberlein, and B. Kennedy, "Replicative and Chronological Aging in Saccharomyces cerevisiae", Cell Metabolism, vol. 16, pp. 18-31, 2012. http://dx.doi.org/10.1016/j.cmet.2012.06.002
- L. Váchová, M. Čáp, and Z. Palková, "Yeast Colonies: A Model for Studies of Aging, Environmental Adaptation, and Longevity", Oxidative Medicine and Cellular Longevity, vol. 2012, pp. 1-8, 2012. http://dx.doi.org/10.1155/2012/601836
- A. Denoth Lippuner, T. Julou, and Y. Barral, "Budding yeast as a model organism to study the effects of age", FEMS Microbiology Reviews, vol. 38, pp. 300-325, 2014. http://dx.doi.org/10.1111/1574-6976.12060
- T. Nyström, and B. Liu, "Protein quality control in time and space - links to cellular aging", FEMS Yeast Research, vol. 14, pp. 40-48, 2013. http://dx.doi.org/10.1111/1567-1364.12095
- J. „Weissman, C. Guthrie, G. Fink, and . editors“, "„Guide", „Academic, 0.
- D. Botstein, and G.R. Fink, "Yeast: An Experimental Organism for 21st Century Biology", Genetics, vol. 189, pp. 695-704, 2011. http://dx.doi.org/10.1534/genetics.111.130765
- S.S. Lee, I.A. Vizcarra, D.H.E.W. Huberts, L.P. Lee, and M. Heinemann, "Whole lifespan microscopic observation of budding yeast aging through a microfluidic dissection platform", Proceedings of the National Academy of Sciences, vol. 109, pp. 4916-4920, 2012. http://dx.doi.org/10.1073/pnas.1113505109
- G.L. Sutphin, B.A. Olsen, B.K. Kennedy, and M. Kaeberlein, "Genome-Wide Analysis of Yeast Aging", Subcellular Biochemistry, pp. 251-289, 2011. http://dx.doi.org/10.1007/978-94-007-2561-4_12
- Z. Xie, Y. Zhang, K. Zou, O. Brandman, C. Luo, Q. Ouyang, and H. Li, "Molecular phenotyping of aging in single yeast cells using a novel microfluidic device", Aging Cell, vol. 11, pp. 599-606, 2012. http://dx.doi.org/10.1111/j.1474-9726.2012.00821.x
- Y. Zhang, C. Luo, K. Zou, Z. Xie, O. Brandman, Q. Ouyang, and H. Li, "Single Cell Analysis of Yeast Replicative Aging Using a New Generation of Microfluidic Device", PLoS ONE, vol. 7, pp. e48275, 2012. http://dx.doi.org/10.1371/journal.pone.0048275
- T. Eisenberg, H. Knauer, A. Schauer, S. Büttner, C. Ruckenstuhl, D. Carmona-Gutierrez, J. Ring, S. Schroeder, C. Magnes, L. Antonacci, H. Fussi, L. Deszcz, R. Hartl, E. Schraml, A. Criollo, E. Megalou, D. Weiskopf, P. Laun, G. Heeren, M. Breitenbach, B. Grubeck-Loebenstein, E. Herker, B. Fahrenkrog, K. Fröhlich, F. Sinner, N. Tavernarakis, N. Minois, G. Kroemer, and F. Madeo, "Induction of autophagy by spermidine promotes longevity", Nature Cell Biology, vol. 11, pp. 1305-1314, 2009. http://dx.doi.org/10.1038/ncb1975
- A.A. Goldberg, V.R. Richard, P. Kyryakov, S.D. Bourque, A. Beach, M.T. Burstein, A. Glebov, O. Koupaki, T. Boukh-Viner, C. Gregg, M. Juneau, A.M. English, D.Y. Thomas, and V.I. Titorenko, "Chemical genetic screen identifies lithocholic acid as an anti-aging compound that extends yeast chronological life span in a TOR-independent manner, by modulating housekeeping longevity assurance processes.", Aging, 2010. http://www.ncbi.nlm.nih.gov/pubmed/20622262
- P. Kapahi, D. Chen, A.N. Rogers, S.D. Katewa, P.W. Li, E.L. Thomas, and L. Kockel, "With TOR, Less Is More: A Key Role for the Conserved Nutrient-Sensing TOR Pathway in Aging", Cell Metabolism, vol. 11, pp. 453-465, 2010. http://dx.doi.org/10.1016/j.cmet.2010.05.001
- D.S. Evans, P. Kapahi, W. Hsueh, and L. Kockel, "TOR signaling never gets old: Aging, longevity and TORC1 activity", Ageing Research Reviews, vol. 10, pp. 225-237, 2011. http://dx.doi.org/10.1016/j.arr.2010.04.001
- N. Minois, D. Carmona-Gutierrez, and F. Madeo, "Polyamines in aging and disease.", Aging, 2011. http://www.ncbi.nlm.nih.gov/pubmed/21869457
- S.M. Jazwinski, "The Retrograde Response and Other Pathways of Interorganelle Communication in Yeast Replicative Aging", Subcellular Biochemistry, pp. 79-100, 2011. http://dx.doi.org/10.1007/978-94-007-2561-4_4
- S.M. Jazwinski, "The retrograde response: When mitochondrial quality control is not enough", Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, vol. 1833, pp. 400-409, 2013. http://dx.doi.org/10.1016/j.bbamcr.2012.02.010
- A. Leonov, and V.I. Titorenko, "A network of interorganellar communications underlies cellular aging", IUBMB Life, vol. 65, pp. 665-674, 2013. http://dx.doi.org/10.1002/iub.1183
- B.P. Hubbard, and D.A. Sinclair, "Small molecule SIRT1 activators for the treatment of aging and age-related diseases", Trends in Pharmacological Sciences, vol. 35, pp. 146-154, 2014. http://dx.doi.org/10.1016/j.tips.2013.12.004
- D.A. Sinclair, and L. Guarente, "Small-Molecule Allosteric Activators of Sirtuins", Annual Review of Pharmacology and Toxicology, vol. 54, pp. 363-380, 2014. http://dx.doi.org/10.1146/annurev-pharmtox-010611-134657
- A.A. Goldberg, S.D. Bourque, P. Kyryakov, C. Gregg, T. Boukh-Viner, A. Beach, M.T. Burstein, G. Machkalyan, V. Richard, S. Rampersad, D. Cyr, S. Milijevic, and V.I. Titorenko, "Effect of calorie restriction on the metabolic history of chronologically aging yeast", Experimental Gerontology, vol. 44, pp. 555-571, 2009. http://dx.doi.org/10.1016/j.exger.2009.06.001
- K.K. Steffen, B.K. Kennedy, and M. Kaeberlein, "Measuring Replicative Life Span in the Budding Yeast", Journal of Visualized Experiments, 2009. http://dx.doi.org/10.3791/1209
- J. Hu, M. Wei, M.G. Mirisola, and V.D. Longo, "Assessing Chronological Aging in Saccharomyces cerevisiae", Methods in Molecular Biology, pp. 463-472, 2012. http://dx.doi.org/10.1007/978-1-62703-239-1_30
- D.A. Sinclair, "Studying the Replicative Life Span of Yeast Cells", Methods in Molecular Biology, pp. 49-63, 2013. http://dx.doi.org/10.1007/978-1-62703-556-9_5
- C.R. Burtner, C.J. Murakami, and M. Kaeberlein, "A Genomic Approach to Yeast Chronological Aging", Methods in Molecular Biology, pp. 101-114, 2009. http://dx.doi.org/10.1007/978-1-59745-540-4_6
- C. Murakami, and M. Kaeberlein, "Quantifying Yeast Chronological Life Span by Outgrowth of Aged Cells", Journal of Visualized Experiments, 2009. http://dx.doi.org/10.3791/1156
- Z. Wu, L. Song, S.Q. Liu, and D. Huang, "A high throughput screening assay for determination of chronological lifespan of yeast", Experimental Gerontology, vol. 46, pp. 915-922, 2011. http://dx.doi.org/10.1016/j.exger.2011.08.002
- P. Fabrizio, and V.D. Longo, "The Chronological Life Span of Saccharomyces cerevisiae", Methods in Molecular Biology, pp. 89-95, 2007. http://dx.doi.org/10.1007/978-1-59745-361-5_8
- P.W. Piper, "Maximising the Yeast Chronological Lifespan", Subcellular Biochemistry, pp. 145-159, 2011. http://dx.doi.org/10.1007/978-94-007-2561-4_7
- V.D. Longo, and B.K. Kennedy, "Sirtuins in Aging and Age-Related Disease", Cell, vol. 126, pp. 257-268, 2006. http://dx.doi.org/10.1016/j.cell.2006.07.002
- V.D. Longo, and P. Fabrizio, "Chronological Aging in Saccharomyces cerevisiae", Subcellular Biochemistry, pp. 101-121, 2011. http://dx.doi.org/10.1007/978-94-007-2561-4_5
- K.J. Bitterman, O. Medvedik, and D.A. Sinclair, "Longevity Regulation inSaccharomyces cerevisiae: Linking Metabolism, Genome Stability, and Heterochromatin", Microbiology and Molecular Biology Reviews, vol. 67, pp. 376-399, 2003. http://dx.doi.org/10.1128/MMBR.67.3.376-399.2003
- K. Steinkraus, M. Kaeberlein, and B. Kennedy, "Replicative Aging in Yeast: The Means to the End", Annual Review of Cell and Developmental Biology, vol. 24, pp. 29-54, 2008. http://dx.doi.org/10.1146/annurev.cellbio.23.090506.123509
- V. Št’ovíček, L. Váchová, M. Kuthan, and Z. Palková, "General factors important for the formation of structured biofilm-like yeast colonies", Fungal Genetics and Biology, vol. 47, pp. 1012-1022, 2010. http://dx.doi.org/10.1016/j.fgb.2010.08.005
- L. Váchová, and Z. Palková, "Aging and longevity of yeast colony populations: metabolic adaptation and differentiation", Biochemical Society Transactions, vol. 39, pp. 1471-1475, 2011. http://dx.doi.org/10.1042/BST0391471
- M. Čáp, L. Štěpánek, K. Harant, L. Váchová, and Z. Palková, "Cell Differentiation within a Yeast Colony: Metabolic and Regulatory Parallels with a Tumor-Affected Organism", Molecular Cell, vol. 46, pp. 436-448, 2012. http://dx.doi.org/10.1016/j.molcel.2012.04.001
- M. Čáp, L. Váchová, and Z. Palková, "Reactive Oxygen Species in the Signaling and Adaptation of Multicellular Microbial Communities", Oxidative Medicine and Cellular Longevity, vol. 2012, pp. 1-13, 2012. http://dx.doi.org/10.1155/2012/976753
- C. Mazzoni, E. Mangiapelo, V. Palermo, and C. Falcone, "Hypothesis: is yeast a clock model to study the onset of humans aging phenotypes?", Frontiers in Oncology, vol. 2, 2012. http://dx.doi.org/10.3389/fonc.2012.00203
- L. Váchová, L. Hatáková, M. Čáp, M. Pokorná, and Z. Palková, "Rapidly Developing Yeast Microcolonies Differentiate in a Similar Way to Aging Giant Colonies", Oxidative Medicine and Cellular Longevity, vol. 2013, pp. 1-9, 2013. http://dx.doi.org/10.1155/2013/102485
- Z. Palková, D. Wilkinson, and L. Váchová, "Aging and differentiation in yeast populations: elders with different properties and functions", FEMS Yeast Research, vol. 14, pp. 96-108, 2013. http://dx.doi.org/10.1111/1567-1364.12103
- V. Šťovíček, L. Váchová, M. Begany, D. Wilkinson, and Z. Palková, "Global changes in gene expression associated with phenotypic switching of wild yeast", BMC Genomics, vol. 15, 2014. http://dx.doi.org/10.1186/1471-2164-15-136
- K.T. Howitz, and D.A. Sinclair, "Xenohormesis: Sensing the Chemical Cues of Other Species", Cell, vol. 133, pp. 387-391, 2008. http://dx.doi.org/10.1016/j.cell.2008.04.019
- A.A. Goldberg, P. Kyryakov, S.D. Bourque, and V.I. Titorenko, "Xenohormetic, hormetic and cytostatic selective forces driving longevity at the ecosystemic level.", Aging, 2010. http://www.ncbi.nlm.nih.gov/pubmed/20693605
- M.T. Burstein, A. Beach, V.R. Richard, O. Koupaki, A. Gomez-Perez, A.A. Goldberg, P. Kyryakov, S.D. Bourque, A. Glebov, and V.I. Titorenko, "Interspecies Chemical Signals Released into the Environment may Create Xenohormetic, Hormetic and Cytostatic Selective Forces that Drive the Ecosystemic Evolution of Longevity Regulation Mechanisms", Dose-Response, vol. 10, 2011. http://dx.doi.org/10.2203/dose-response.11-011.Titorenko
- K.T. Howitz, K.J. Bitterman, H.Y. Cohen, D.W. Lamming, S. Lavu, J.G. Wood, R.E. Zipkin, P. Chung, A. Kisielewski, L. Zhang, B. Scherer, and D.A. Sinclair, "Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan", Nature, vol. 425, pp. 191-196, 2003. http://dx.doi.org/10.1038/nature01960
- D.W. Lamming, J.G. Wood, and D.A. Sinclair, "MicroReview: Small molecules that regulate lifespan: evidence for xenohormesis", Molecular Microbiology, vol. 53, pp. 1003-1009, 2004. http://dx.doi.org/10.1111/j.1365-2958.2004.04209.x
- N.D. Bonawitz, and G.S. Shadel, "Rethinking the Mitochondrial Theory of Aging: The Role of Mitochondrial Gene Expression in Lifespan Determination", Cell Cycle, vol. 6, pp. 1574-1578, 2007. http://dx.doi.org/10.4161/cc.6.13.4457
- N.D. Bonawitz, M. Chatenay-Lapointe, Y. Pan, and G.S. Shadel, "Reduced TOR Signaling Extends Chronological Life Span via Increased Respiration and Upregulation of Mitochondrial Gene Expression", Cell Metabolism, vol. 5, pp. 265-277, 2007. http://dx.doi.org/10.1016/j.cmet.2007.02.009
- Y. Pan, and G.S. Shadel, "Extension of chronological life span by reduced TOR signaling requires down-regulation of Sch9p and involves increased mitochondrial OXPHOS complex density.", Aging, 2009. http://www.ncbi.nlm.nih.gov/pubmed/20157595
- M. Wei, P. Fabrizio, F. Madia, J. Hu, H. Ge, L.M. Li, and V.D. Longo, "Tor1/Sch9-Regulated Carbon Source Substitution Is as Effective as Calorie Restriction in Life Span Extension", PLoS Genetics, vol. 5, pp. e1000467, 2009. http://dx.doi.org/10.1371/journal.pgen.1000467
- A. Mesquita, M. Weinberger, A. Silva, B. Sampaio-Marques, B. Almeida, C. Leão, V. Costa, F. Rodrigues, W.C. Burhans, and P. Ludovico, "Caloric restriction or catalase inactivation extends yeast chronological lifespan by inducing H 2 O 2 and superoxide dismutase activity", Proceedings of the National Academy of Sciences, vol. 107, pp. 15123-15128, 2010. http://dx.doi.org/10.1073/pnas.1004432107
- A. Beach, and V.I. Titorenko, "In search of housekeeping pathways that regulate longevity", Cell Cycle, vol. 10, pp. 3042-3044, 2011. http://dx.doi.org/10.4161/cc.10.18.16947
- Y. Pan, "Mitochondria, reactive oxygen species, and chronological aging: A message from yeast", Experimental Gerontology, vol. 46, pp. 847-852, 2011. http://dx.doi.org/10.1016/j.exger.2011.08.007
- Y. Pan, E. Schroeder, A. Ocampo, A. Barrientos, and G. Shadel, "Regulation of Yeast Chronological Life Span by TORC1 via Adaptive Mitochondrial ROS Signaling", Cell Metabolism, vol. 13, pp. 668-678, 2011. http://dx.doi.org/10.1016/j.cmet.2011.03.018
- V.I. Titorenko, and S.R. Terlecky, "Peroxisome Metabolism and Cellular Aging", Traffic, vol. 12, pp. 252-259, 2010. http://dx.doi.org/10.1111/j.1600-0854.2010.01144.x
- A. Beach, M.T. Burstein, V.R. Richard, A. Leonov, S. Levy, and V.I. Titorenko, "Integration of peroxisomes into an endomembrane system that governs cellular aging", Frontiers in Physiology, vol. 3, 2012. http://dx.doi.org/10.3389/fphys.2012.00283
- M.T. Burstein, P. Kyryakov, A. Beach, V.R. Richard, O. Koupaki, A. Gomez-Perez, A. Leonov, S. Levy, F. Noohi, and V.I. Titorenko, "Lithocholic acid extends longevity of chronologically aging yeast only if added at certain critical periods of their lifespan", Cell Cycle, vol. 11, pp. 3443-3462, 2012. http://dx.doi.org/10.4161/cc.21754
- L. Cai, and B.P. Tu, "Driving the Cell Cycle Through Metabolism", Annual Review of Cell and Developmental Biology, vol. 28, pp. 59-87, 2012. http://dx.doi.org/10.1146/annurev-cellbio-092910-154010
- P. Kyryakov, A. Beach, V.R. Richard, M.T. Burstein, A. Leonov, S. Levy, and V.I. Titorenko, "Caloric Restriction Extends Yeast Chronological Lifespan by Altering a Pattern of Age-Related Changes in Trehalose Concentration", Frontiers in Physiology, vol. 3, 2012. http://dx.doi.org/10.3389/fphys.2012.00256
- A. Ocampo, J. Liu, E. Schroeder, G. Shadel, and A. Barrientos, "Mitochondrial Respiratory Thresholds Regulate Yeast Chronological Life Span and its Extension by Caloric Restriction", Cell Metabolism, vol. 16, pp. 55-67, 2012. http://dx.doi.org/10.1016/j.cmet.2012.05.013
- Y. Barral, "A new answer to old questions", eLife, vol. 2, 2013. http://dx.doi.org/10.7554/eLife.00515
- A. Beach, V.R. Richard, A. Leonov, M.T. Burstein, S.D. Bourque, O. Koupaki, M. Juneau, R. Feldman, T. Iouk, and V.I. Titorenko, "Mitochondrial membrane lipidome defines yeast longevity.", Aging, 2013. http://www.ncbi.nlm.nih.gov/pubmed/23924582
- A. Beach, and V.I. Titorenko, "Essential Roles of Peroxisomally Produced and Metabolized Biomolecules in Regulating Yeast Longevity", Subcellular Biochemistry, pp. 153-167, 2013. http://dx.doi.org/10.1007/978-94-007-6889-5_9
- N. Brandes, H. Tienson, A. Lindemann, V. Vitvitsky, D. Reichmann, R. Banerjee, and U. Jakob, "Time line of redox events in aging postmitotic cells", eLife, vol. 2, 2013. http://dx.doi.org/10.7554/eLife.00306
- M. Hachinohe, M. Yamane, D. Akazawa, K. Ohsawa, M. Ohno, Y. Terashita, and H. Masumoto, "A Reduction in Age-Enhanced Gluconeogenesis Extends Lifespan", PLoS ONE, vol. 8, pp. e54011, 2013. http://dx.doi.org/10.1371/journal.pone.0054011
- M. Mirisola, and V. Longo, "A Radical Signal Activates the Epigenetic Regulation of Longevity", Cell Metabolism, vol. 17, pp. 812-813, 2013. http://dx.doi.org/10.1016/j.cmet.2013.05.015
- I. Orlandi, R. Ronzulli, N. Casatta, and M. Vai, "Ethanol and Acetate Acting as Carbon/Energy Sources Negatively Affect Yeast Chronological Aging", Oxidative Medicine and Cellular Longevity, vol. 2013, pp. 1-10, 2013. http://dx.doi.org/10.1155/2013/802870
- V.R. Richard, A. Leonov, A. Beach, M.T. Burstein, O. Koupaki, A. Gomez-Perez, S. Levy, L. Pluska, S. Mattie, R. Rafesh, T. Iouk, S. Sheibani, M. Greenwood, H. Vali, and V.I. Titorenko, "Macromitophagy is a longevity assurance process that in chronologically aging yeast limited in calorie supply sustains functional mitochondria and maintains cellular lipid homeostasis.", Aging, 2013. http://www.ncbi.nlm.nih.gov/pubmed/23553280
- E. Schroeder, N. Raimundo, and G. Shadel, "Epigenetic Silencing Mediates Mitochondria Stress-Induced Longevity", Cell Metabolism, vol. 17, pp. 954-964, 2013. http://dx.doi.org/10.1016/j.cmet.2013.04.003
- E.B. Tahara, F.M. Cunha, T.O. Basso, B.E. Della Bianca, A.K. Gombert, and A.J. Kowaltowski, "Calorie Restriction Hysteretically Primes Aging Saccharomyces cerevisiae toward More Effective Oxidative Metabolism", PLoS ONE, vol. 8, pp. e56388, 2013. http://dx.doi.org/10.1371/journal.pone.0056388
- D. Martins, V.I. Titorenko, and A.M. English, "Cells with Impaired Mitochondrial H2O2Sensing Generate Less•OH Radicals and Live Longer", Antioxidants & Redox Signaling, vol. 21, pp. 1490-1503, 2014. http://dx.doi.org/10.1089/ars.2013.5575
- B. Chen, M. Retzlaff, T. Roos, and J. Frydman, "Cellular Strategies of Protein Quality Control", Cold Spring Harbor Perspectives in Biology, vol. 3, pp. a004374-a004374, 2011. http://dx.doi.org/10.1101/cshperspect.a004374
- S.L. Lindquist, and J.W. Kelly, "Chemical and Biological Approaches for Adapting Proteostasis to Ameliorate Protein Misfolding and Aggregation Diseases-Progress and Prognosis", Cold Spring Harbor Perspectives in Biology, vol. 3, pp. a004507-a004507, 2011. http://dx.doi.org/10.1101/cshperspect.a004507
- R.C. Taylor, and A. Dillin, "Aging as an Event of Proteostasis Collapse", Cold Spring Harbor Perspectives in Biology, vol. 3, pp. a004440-a004440, 2011. http://dx.doi.org/10.1101/cshperspect.a004440
- Y.E. Kim, M.S. Hipp, A. Bracher, M. Hayer-Hartl, and F. Ulrich Hartl, "Molecular Chaperone Functions in Protein Folding and Proteostasis", Annual Review of Biochemistry, vol. 82, pp. 323-355, 2013. http://dx.doi.org/10.1146/annurev-biochem-060208-092442
- J.E. Legakis, J.I. Koepke, C. Jedeszko, F. Barlaskar, L.J. Terlecky, H.J. Edwards, P.A. Walton, and S.R. Terlecky, "Peroxisome Senescence in Human Fibroblasts", Molecular Biology of the Cell, vol. 13, pp. 4243-4255, 2002. http://dx.doi.org/10.1091/mbc.E02-06-0322
- S.R. Terlecky, J.I. Koepke, and P.A. Walton, "Peroxisomes and aging", Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, vol. 1763, pp. 1749-1754, 2006. http://dx.doi.org/10.1016/j.bbamcr.2006.08.017
- C. Ma, G. Agrawal, and S. Subramani, "Peroxisome assembly: matrix and membrane protein biogenesis", Journal of Cell Biology, vol. 193, pp. 7-16, 2011. http://dx.doi.org/10.1083/jcb.201010022
- X. Liu, C. Ma, and S. Subramani, "Recent advances in peroxisomal matrix protein import", Current Opinion in Cell Biology, vol. 24, pp. 484-489, 2012. http://dx.doi.org/10.1016/j.ceb.2012.05.003
- S. Hasan, H.W. Platta, and R. Erdmann, "Import of proteins into the peroxisomal matrix", Frontiers in Physiology, vol. 4, 2013. http://dx.doi.org/10.3389/fphys.2013.00261
- O. Ivashchenko, P.P. Van Veldhoven, C. Brees, Y. Ho, S.R. Terlecky, and M. Fransen, "Intraperoxisomal redox balance in mammalian cells: oxidative stress and interorganellar cross-talk", Molecular Biology of the Cell, vol. 22, pp. 1440-1451, 2011. http://dx.doi.org/10.1091/mbc.E10-11-0919
- M. Islinger, S. Grille, H.D. Fahimi, and M. Schrader, "The peroxisome: an update on mysteries", Histochemistry and Cell Biology, vol. 137, pp. 547-574, 2012. http://dx.doi.org/10.1007/s00418-012-0941-4
- P.A. Walton, "Effects of peroxisomal catalase inhibition on mitochondrial function", Frontiers in Physiology, vol. 3, 2012. http://dx.doi.org/10.3389/fphys.2012.00108
- B. Wang, P.P. Van Veldhoven, C. Brees, N. Rubio, M. Nordgren, O. Apanasets, M. Kunze, M. Baes, P. Agostinis, and M. Fransen, "Mitochondria are targets for peroxisome-derived oxidative stress in cultured mammalian cells", Free Radical Biology and Medicine, vol. 65, pp. 882-894, 2013. http://dx.doi.org/10.1016/j.freeradbiomed.2013.08.173
- M. Nordgren, and M. Fransen, "Peroxisomal metabolism and oxidative stress", Biochimie, vol. 98, pp. 56-62, 2014. http://dx.doi.org/10.1016/j.biochi.2013.07.026
- V.I. Titorenko, and R.A. Rachubinski, "The peroxisome", The Journal of Cell Biology, vol. 164, pp. 641-645, 2004. http://dx.doi.org/10.1083/jcb.200312081
- A. Goldberg, S. Bourque, P. Kyryakov, T. Boukh-Viner, C. Gregg, A. Beach, M. Burstein, G. Machkalyan, V. Richard, S. Rampersad, and V. Titorenko, "A novel function of lipid droplets in regulating longevity", Biochemical Society Transactions, vol. 37, pp. 1050-1055, 2009. http://dx.doi.org/10.1042/BST0371050
- J.K. Hiltunen, A.M. Mursula, H. Rottensteiner, R.K. Wierenga, A.J. Kastaniotis, and A. Gurvitz, "The biochemistry of peroxisomal beta-oxidation in the yeast Saccharomyces cerevisiae.", FEMS microbiology reviews, 2003. http://www.ncbi.nlm.nih.gov/pubmed/12697341
- A. Kawałek, S.D. Lefevre, M. Veenhuis, and I.J. van der Klei, "Peroxisomal catalase deficiency modulates yeast lifespan depending on growth conditions.", Aging, 2013. http://www.ncbi.nlm.nih.gov/pubmed/23425686
- S.D. Lefevre, C.W. Roermund, R.J.A. Wanders, M. Veenhuis, and I.J. Klei, "The significance of peroxisome function in chronological aging ofSaccharomyces cerevisiae", Aging Cell, vol. 12, pp. 784-793, 2013. http://dx.doi.org/10.1111/acel.12113
- N. Minois, "Molecular basis of the 'anti-aging' effect of spermidine and other natural polyamines - a mini-review.", Gerontology, 2014. http://www.ncbi.nlm.nih.gov/pubmed/24481223
- E. Morselli, L. Galluzzi, O. Kepp, A. Criollo, M.C. Maiuri, N. Tavernarakis, F. Madeo, and G. Kroemer, "Autophagy mediates pharmacological lifespan extension by spermidine and resveratrol.", Aging, 2009. http://www.ncbi.nlm.nih.gov/pubmed/20157579
- E. Morselli, G. Mariño, M.V. Bennetzen, T. Eisenberg, E. Megalou, S. Schroeder, S. Cabrera, P. Bénit, P. Rustin, A. Criollo, O. Kepp, L. Galluzzi, S. Shen, S.A. Malik, M.C. Maiuri, Y. Horio, C. López-Otín, J.S. Andersen, N. Tavernarakis, F. Madeo, and G. Kroemer, "Spermidine and resveratrol induce autophagy by distinct pathways converging on the acetylproteome", Journal of Cell Biology, vol. 192, pp. 615-629, 2011. http://dx.doi.org/10.1083/jcb.201008167
- D. Binns, T. Januszewski, Y. Chen, J. Hill, V.S. Markin, Y. Zhao, C. Gilpin, K.D. Chapman, R.G. Anderson, and J.M. Goodman, "An intimate collaboration between peroxisomes and lipid bodies", The Journal of Cell Biology, vol. 173, pp. 719-731, 2006. http://dx.doi.org/10.1083/jcb.200511125
- B. D'Autréaux, and M.B. Toledano, "ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis", Nature Reviews Molecular Cell Biology, vol. 8, pp. 813-824, 2007. http://dx.doi.org/10.1038/nrm2256
- M. Giorgio, M. Trinei, E. Migliaccio, and P.G. Pelicci, "Hydrogen peroxide: a metabolic by-product or a common mediator of ageing signals?", Nature Reviews Molecular Cell Biology, vol. 8, pp. 722-728, 2007. http://dx.doi.org/10.1038/nrm2240
- E.A. Veal, A.M. Day, and B.A. Morgan, "Hydrogen Peroxide Sensing and Signaling", Molecular Cell, vol. 26, pp. 1-14, 2007. http://dx.doi.org/10.1016/j.molcel.2007.03.016
- J.M. Goodman, "The Gregarious Lipid Droplet", Journal of Biological Chemistry, vol. 283, pp. 28005-28009, 2008. http://dx.doi.org/10.1074/jbc.R800042200
- O. Adeyo, P.J. Horn, S. Lee, D.D. Binns, A. Chandrahas, K.D. Chapman, and J.M. Goodman, "The yeast lipin orthologue Pah1p is important for biogenesis of lipid droplets", Journal of Cell Biology, vol. 192, pp. 1043-1055, 2011. http://dx.doi.org/10.1083/jcb.201010111
- S.D. Kohlwein, M. Veenhuis, and I.J. van der Klei, "Lipid Droplets and Peroxisomes: Key Players in Cellular Lipid Homeostasis or A Matter of Fat—Store ’em Up or Burn ’em Down", Genetics, vol. 193, pp. 1-50, 2013. http://dx.doi.org/10.1534/genetics.112.143362
- S. Sheibani, V. Richard, A. Beach, A. Leonov, R. Feldman, S. Mattie, L. Khelghatybana, A. Piano, M. Greenwood, H. Vali, and V.I. Titorenko, "Macromitophagy, neutral lipids synthesis, and peroxisomal fatty acid oxidation protect yeast from “liponecrosis”, a previously unknown form of programmed cell death", Cell Cycle, vol. 13, pp. 138-147, 2013. http://dx.doi.org/10.4161/cc.26885
- I.J. van der Klei, H. Yurimoto, Y. Sakai, and M. Veenhuis, "The significance of peroxisomes in methanol metabolism in methylotrophic yeast", Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, vol. 1763, pp. 1453-1462, 2006. http://dx.doi.org/10.1016/j.bbamcr.2006.07.016
- P.W. Piper, N.L. Harris, and M. MacLean, "Preadaptation to efficient respiratory maintenance is essential both for maximal longevity and the retention of replicative potential in chronologically ageing yeast", Mechanisms of Ageing and Development, vol. 127, pp. 733-740, 2006. http://dx.doi.org/10.1016/j.mad.2006.05.004
- H. Lavoie, and M. Whiteway, "Increased Respiration in the sch9Δ Mutant Is Required for Increasing Chronological Life Span but Not Replicative Life Span", Eukaryotic Cell, vol. 7, pp. 1127-1135, 2008. http://dx.doi.org/10.1128/EC.00330-07
- M.T. Burstein, and V.I. Titorenko, "A mitochondrially targeted compound delays aging in yeast through a mechanism linking mitochondrial membrane lipid metabolism to mitochondrial redox biology", Redox Biology, vol. 2, pp. 305-307, 2014. http://dx.doi.org/10.1016/j.redox.2014.01.011
- E.A. Schroeder, and G.S. Shadel, "Crosstalk between mitochondrial stress signals regulates yeast chronological lifespan", Mechanisms of Ageing and Development, vol. 135, pp. 41-49, 2014. http://dx.doi.org/10.1016/j.mad.2013.12.002
- J.R. Broach, "Nutritional Control of Growth and Development in Yeast", Genetics, vol. 192, pp. 73-105, 2012. http://dx.doi.org/10.1534/genetics.111.135731
- C. De Virgilio, "The essence of yeast quiescence", FEMS Microbiology Reviews, vol. 36, pp. 306-339, 2012. http://dx.doi.org/10.1111/j.1574-6976.2011.00287.x
- J. Orzechowski Westholm, S. Tronnersjö, N. Nordberg, I. Olsson, J. Komorowski, and H. Ronne, "Gis1 and Rph1 Regulate Glycerol and Acetate Metabolism in Glucose Depleted Yeast Cells", PLoS ONE, vol. 7, pp. e31577, 2012. http://dx.doi.org/10.1371/journal.pone.0031577
- P. Fabrizio, C. Gattazzo, L. Battistella, M. Wei, C. Cheng, K. McGrew, and V.D. Longo, "Sir2 Blocks Extreme Life-Span Extension", Cell, vol. 123, pp. 655-667, 2005. http://dx.doi.org/10.1016/j.cell.2005.08.042
- C.R. Burtner, C.J. Murakami, B.K. Kennedy, and M. Kaeberlein, "A molecular mechanism of chronological aging in yeast", Cell Cycle, vol. 8, pp. 1256-1270, 2009. http://dx.doi.org/10.4161/cc.8.8.8287
- C.R. Burtner, C.J. Murakami, B. Olsen, B.K. Kennedy, and M. Kaeberlein, "A genomic analysis of chronological longevity factors in budding yeast", Cell Cycle, vol. 10, pp. 1385-1396, 2011. http://dx.doi.org/10.4161/cc.10.9.15464
- C. Murakami, J.R. Delaney, A. Chou, D. Carr, J. Schleit, G.L. Sutphin, E.H. An, A.S. Castanza, M. Fletcher, S. Goswami, S. Higgins, M. Holmberg, J. Hui, M. Jelic, K. Jeong, J.R. Kim, S. Klum, E. Liao, M.S. Lin, W. Lo, H. Miller, R. Moller, Z.J. Peng, T. Pollard, P. Pradeep, D. Pruett, D. Rai, V. Ros, A. Schuster, M. Singh, B.L. Spector, H. Vander Wende, A.M. Wang, B.M. Wasko, B. Olsen, and M. Kaeberlein, "pH neutralization protects against reduction in replicative lifespan following chronological aging in yeast", Cell Cycle, vol. 11, pp. 3087-3096, 2012. http://dx.doi.org/10.4161/cc.21465
- T. Eisenberg, S. Schroeder, A. Andryushkova, T. Pendl, V. Küttner, A. Bhukel, G. Mariño, F. Pietrocola, A. Harger, A. Zimmermann, T. Moustafa, A. Sprenger, E. Jany, S. Büttner, D. Carmona-Gutierrez, C. Ruckenstuhl, J. Ring, W. Reichelt, K. Schimmel, T. Leeb, C. Moser, S. Schatz, L. Kamolz, C. Magnes, F. Sinner, S. Sedej, K. Fröhlich, G. Juhasz, T. Pieber, J. Dengjel, S.J. Sigrist, G. Kroemer, and F. Madeo, "Nucleocytosolic Depletion of the Energy Metabolite Acetyl-Coenzyme A Stimulates Autophagy and Prolongs Lifespan", Cell Metabolism, vol. 19, pp. 431-444, 2014. http://dx.doi.org/10.1016/j.cmet.2014.02.010
- J. Hu, M. Wei, H. Mirzaei, F. Madia, M. Mirisola, C. Amparo, S. Chagoury, B. Kennedy, and V.D. Longo, "Tor‐Sch9 deficiency activates catabolism of the ketone body‐like acetic acid to promote trehalose accumulation and longevity", Aging Cell, vol. 13, pp. 457-467, 2014. http://dx.doi.org/10.1111/acel.12202
- D. Fraenkel, "Yeast intermediary metabolism.", Cold Spring Harbor Laboratory Press, Cold Spring Harbor, 2011.
- J.L. Crespo, T. Powers, B. Fowler, and M.N. Hall, "The TOR-controlled transcription activators GLN3, RTG1, and RTG3 are regulated in response to intracellular levels of glutamine", Proceedings of the National Academy of Sciences, vol. 99, pp. 6784-6789, 2002. http://dx.doi.org/10.1073/pnas.102687599
- R.W. Powers, M. Kaeberlein, S.D. Caldwell, B.K. Kennedy, and S. Fields, "Extension of chronological life span in yeast by decreased TOR pathway signaling", Genes & Development, vol. 20, pp. 174-184, 2006. http://dx.doi.org/10.1101/gad.1381406
- J.L. Jewell, R.C. Russell, and K. Guan, "Amino acid signalling upstream of mTOR", Nature Reviews Molecular Cell Biology, vol. 14, pp. 133-139, 2013. http://dx.doi.org/10.1038/nrm3522
- M. Conrad, J. Schothorst, H.N. Kankipati, G. Van Zeebroeck, M. Rubio-Texeira, and J.M. Thevelein, "Nutrient sensing and signaling in the yeastSaccharomyces cerevisiae", FEMS Microbiology Reviews, vol. 38, pp. 254-299, 2014. http://dx.doi.org/10.1111/1574-6976.12065
- M. Shimobayashi, and M.N. Hall, "Making new contacts: the mTOR network in metabolism and signalling crosstalk", Nature Reviews Molecular Cell Biology, vol. 15, pp. 155-162, 2014. http://dx.doi.org/10.1038/nrm3757
- E. Swinnen, R. Ghillebert, T. Wilms, and J. Winderickx, "Molecular mechanisms linking the evolutionary conserved TORC1-Sch9 nutrient signalling branch to lifespan regulation inSaccharomyces cerevisiae", FEMS Yeast Research, vol. 14, pp. 17-32, 2013. http://dx.doi.org/10.1111/1567-1364.12097
- F. Barea, and D. Bonatto, "Aging defined by a chronologic–replicative protein network in Saccharomyces cerevisiae: An interactome analysis", Mechanisms of Ageing and Development, vol. 130, pp. 444-460, 2009. http://dx.doi.org/10.1016/j.mad.2009.04.005
- D.R. Lorenz, C.R. Cantor, and J.J. Collins, "A network biology approach to aging in yeast", Proceedings of the National Academy of Sciences, vol. 106, pp. 1145-1150, 2009. http://dx.doi.org/10.1073/pnas.0812551106
- E. Borklu Yucel, and K.O. Ulgen, "A Network-Based Approach on Elucidating the Multi-Faceted Nature of Chronological Aging in S. cerevisiae", PLoS ONE, vol. 6, pp. e29284, 2011. http://dx.doi.org/10.1371/journal.pone.0029284
- D. Gozuacik, S. Bialik, T. Raveh, G. Mitou, G. Shohat, H. Sabanay, N. Mizushima, T. Yoshimori, and A. Kimchi, "DAP-kinase is a mediator of endoplasmic reticulum stress-induced caspase activation and autophagic cell death", Cell Death & Differentiation, vol. 15, pp. 1875-1886, 2008. http://dx.doi.org/10.1038/cdd.2008.121
- A. Eisenberg-Lerner, S. Bialik, H. Simon, and A. Kimchi, "Life and death partners: apoptosis, autophagy and the cross-talk between them", Cell Death & Differentiation, vol. 16, pp. 966-975, 2009. http://dx.doi.org/10.1038/cdd.2009.33
- S. Bialik, E. Zalckvar, Y. Ber, A.D. Rubinstein, and A. Kimchi, "Systems biology analysis of programmed cell death", Trends in Biochemical Sciences, vol. 35, pp. 556-564, 2010. http://dx.doi.org/10.1016/j.tibs.2010.04.008
- E. Zalckvar, S. Bialik, and A. Kimchi, "The road not taken: a systems level strategy for analyzing the cell death network.", Autophagy, 2010. http://www.ncbi.nlm.nih.gov/pubmed/20574161
- E. Zalckvar, N. Yosef, S. Reef, Y. Ber, A.D. Rubinstein, I. Mor, R. Sharan, E. Ruppin, and A. Kimchi, "A systems level strategy for analyzing the cell death network: implication in exploring the apoptosis/autophagy connection", Cell Death & Differentiation, vol. 17, pp. 1244-1253, 2010. http://dx.doi.org/10.1038/cdd.2010.7
- A.J. Munoz, K. Wanichthanarak, E. Meza, and D. Petranovic, "Systems biology of yeast cell death", FEMS Yeast Research, vol. 12, pp. 249-265, 2012. http://dx.doi.org/10.1111/j.1567-1364.2011.00781.x
- A.D. Rubinstein, and A. Kimchi, "Life in the balance – a mechanistic view of the crosstalk between autophagy and apoptosis", Journal of Cell Science, vol. 125, pp. 5259-5268, 2012. http://dx.doi.org/10.1242/jcs.115865
- M.M. Young, M. Kester, and H. Wang, "Sphingolipids: regulators of crosstalk between apoptosis and autophagy", Journal of Lipid Research, vol. 54, pp. 5-19, 2013. http://dx.doi.org/10.1194/jlr.R031278
- S. Büttner, T. Eisenberg, E. Herker, D. Carmona-Gutierrez, G. Kroemer, and F. Madeo, "Why yeast cells can undergo apoptosis: death in times of peace, love, and war", The Journal of Cell Biology, vol. 175, pp. 521-525, 2006. http://dx.doi.org/10.1083/jcb.200608098
- D. Carmona-Gutierrez, T. Eisenberg, S. Büttner, C. Meisinger, G. Kroemer, and F. Madeo, "Apoptosis in yeast: triggers, pathways, subroutines", Cell Death & Differentiation, vol. 17, pp. 763-773, 2010. http://dx.doi.org/10.1038/cdd.2009.219
ACKNOWLEDGMENTS
V.I.T. research is supported by grants from the NSERC of Canada and Concordia University Chair Fund. V.I.T. is a Concordia University Research Chair in Genomics, Cell Biology and Aging.
COPYRIGHT
© 2014

Cell-autonomous mechanisms of chronological aging in the yeast Saccharomyces cerevisiae by Anthony Arlia-Ciommo et al. is licensed under a Creative Commons Attribution 4.0 International License.